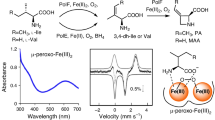
Abstract
Azetidine is a prominent pharmacophore present in dozens of drug-related molecules of both natural and synthetic origins. But how nature builds this moiety has long remained enigmatic. Here we address the full deciphering of a two-metalloenzyme cascade leading to polyoximic acid, an azetidine-containing moiety of the fungicide polyoxin. We demonstrate that the PolE enzyme functions as an Fe2+/pterin-dependent l-isoleucine desaturase. Moreover we illustrate that PolF is a new member of the emerging haem-oxygenase-like diiron oxidases, converting the desaturated l-isoleucine to polyoximic acid via an intramolecular C–N cyclization. Remarkably, we also establish that PolF exhibits dual functionality, orchestrating the sequential desaturation and cyclization with l-isoleucine as the initial substrate. Finally, our combined structural and quantum-mechanics/molecular-mechanics studies show that the PolF enzyme employs an extraordinary mechanism for the construction of the azetidine-containing moiety. These findings expand our knowledge on the catalysis of metalloenzymes and open the way for rational access of more azetidine-related molecules.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The atomic coordinates and structure factors of the reported structures have been deposited in the PDB under accession codes as follows: apo SnPolE, 9J2E; SnPolE–BH4, 9J2M; SnPolE–BH4–l-isoleucine, 9J2P; SnPolEF140A, 9J33; SnPolEY157A, 9J2R; SnPolEY280A, 9J30; apo SmPolE, 9J3G; Fe(II)–SzPolF–l-isoleucine, 9J2C; Fe2(II/II)–SzPolF–l-isoleucine, 9J2B; Fe(II)–SzPolF–2, 9J28; Fe2(II/II)–SzPolF–2, 9J2A; Fe2(II/II)–SzPolF–1, 9J29; Fe2(II/II)–PolF–l-isoleucine, 9J3V; and Fe(II)–SaPolF–l-isoleucine, 9J27. All other data generated and analysed supporting the findings of this study are available within the paper and Supplementary Information. Source data are provided with this paper.
References
Wearing, E. R. et al. Visible light–mediated aza Paternò–Büchi reaction of acyclic oximes and alkenes to azetidines. Science 384, 1468–1476 (2024).
Becker, M. R., Wearing, E. R. & Schindler, C. S. Synthesis of azetidines via visible-light-mediated intermolecular [2+2] photocycloadditions. Nat. Chem. 12, 898–905 (2020).
Brandi, A., Cicchi, S. & Cordero, F. M. Novel syntheses of azetidines and azetidinones. Chem. Rev. 108, 3988–4035 (2008).
Marshall, C. M., Federice, J. G., Bell, C. N., Cox, P. B. & Njardarson, J. T. An update on the nitrogen heterocycle compositions and properties of U.S. FDA-approved pharmaceuticals (2013–2023). J. Med. Chem. 67, 11622–11655 (2024).
Becker, M. R., Richardson, A. D. & Schindler, C. S. Functionalized azetidines via visible light-enabled aza Paternò-Büchi reactions. Nat. Commun. 10, 5095 (2019).
Fowden, L. Azetidine-2-carboxylic acid: a new constituent of plants. Nature 176, 347–348 (1955).
Rabe, P., Kamps, J. J. A. G., Schofield, C. J. & Lohans, C. T. Roles of 2-oxoglutarate oxygenases and isopenicillin N synthase in β-lactam biosynthesis. Nat. Prod. Rep. 35, 735–756 (2018).
Yan, F. et al. Biosynthesis and heterologous production of vioprolides: rational biosynthetic engineering and unprecedented 4‐methylazetidinecarboxylic acid formation. Angew. Chem. Int. Ed. 57, 8754–8759 (2018).
Hong, Z. et al. Azetidine-containing alkaloids produced by a quorum-sensing regulated nonribosomal peptide synthetase pathway in Pseudomonas aeruginosa. Angew. Chem. Int. Ed. 58, 3178–3182 (2019).
Chen, W. et al. Characterization of the polyoxin biosynthetic gene cluster from Streptomyces cacaoi and engineered production of polyoxin H. J. Biol. Chem. 284, 10627–10638 (2009).
Chen, W. et al. An unusual UMP C-5 methylase in nucleoside antibiotic polyoxin biosynthesis. Protein Cell 7, 673–683 (2016).
Lilla, E. A. & Yokoyama, K. Carbon extension in peptidylnucleoside biosynthesis by radical SAM enzymes. Nat. Chem. Biol. 12, 905–907 (2016).
Draelos, M. M., Thanapipatsiri, A., Sucipto, H. & Yokoyama, K. Cryptic phosphorylation in nucleoside natural product biosynthesis. Nat. Chem. Biol. 17, 213–221 (2021).
Qi, J. et al. Deciphering carbamoylpolyoxamic acid biosynthesis reveals unusual acetylation cycle associated with tandem reduction and sequential hydroxylation. Cell Chem. Biol. 23, 935–944 (2016).
Gust, B., Challis, G. L., Fowler, K., Kieser, T. & Chater, K. F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl Acad. Sci. USA 100, 1541–1546 (2003).
Liao, H. J. et al. Insights into the desaturation of cyclopeptin and its C3 epimer catalyzed by a non‐heme iron nnzyme: structural characterization and mechanism elucidation. Angew. Chem. Int. Ed. 57, 1831–1835 (2018).
Iyer, S. R. et al. Direct coordination of pterin to FeII enables neurotransmitter biosynthesis in the pterin-dependent hydroxylases. Proc. Natl Acad. Sci. USA 118, e2022379118 (2021).
Kappock, T. J. & Caradonna, J. P. Pterin-dependent amino acid hydroxylases. Chem. Rev. 96, 2659–2756 (1996).
Manley, O. M. et al. Self-sacrificial tyrosine cleavage by an Fe:Mn oxygenase for the biosynthesis of para-aminobenzoate in Chlamydia trachomatis. Proc. Natl Acad. Sci. USA 119, e2210908119 (2022).
Iqbal, T., Murugan, S., Rajendran, K., Sidhu, J. S. & Das, D. Unraveling the conversion of fatty acids into terminal alkenes by an integral membrane enzyme, UndB. ACS Catal. 13, 15516–15525 (2023).
Makris, T. M., Chakrabarti, M., Münck, E. & Lipscomb, J. D. A family of diiron monooxygenases catalyzing amino acid beta-hydroxylation in antibiotic biosynthesis. Proc. Natl Acad. Sci. USA 107, 15391–15396 (2010).
Zhang, B. et al. Substrate-triggered formation of a peroxo-Fe2(III/III) intermediate during fatty acid decarboxylation by UndA. J. Am. Chem. Soc. 141, 14510–14514 (2019).
Ng, T. L., Rohac, R., Mitchell, A. J., Boal, A. K. & Balskus, E. P. An N-nitrosating metalloenzyme constructs the pharmacophore of streptozotocin. Nature 566, 94–99 (2019).
McBride, M. J. et al. Structure and assembly of the diiron cofactor in the heme-oxygenase-like domain of the N-nitrosourea-producing enzyme SznF. Proc. Natl Acad. Sci. USA 118, e2015931118 (2021).
Li, H. et al. The structural and functional investigation into an unusual nitrile synthase. Nat. Commun. 14, 7425–7434 (2023).
Adak, S. et al. A single diiron enzyme catalyses the oxidative rearrangement of tryptophan to indole nitrile. Nat. Chem. 16, 1989–1998 (2024).
McBride, M. J. et al. A peroxodiiron(III/III) intermediate mediating both N-hydroxylation steps in biosynthesis of the N-nitrosourea pharmacophore of streptozotocin by the multi-domain metalloenzyme SznF. J. Am. Chem. Soc. 142, 11818–11828 (2020).
Manley, O. M. et al. BesC initiates C–C cleavage through a substrate-triggered and reactive diferric-oeroxo intermediate. J. Am. Chem. Soc. 143, 21416–21424 (2021).
McBride, M. J. et al. Substrate-triggered μ-peroxodiiron(III) intermediate in the 4-chloro-l-lysine-fragmenting heme-oxygenase-like diiron oxidase (HDO) BesC: substrate dissociation from, and C4 targeting by, the intermediate. Biochemistry 61, 689–702 (2022).
Adak, S., Calderone, L. A., Krueger, A., Pandelia, M.-E. & Moore, B. S. Single-enzyme conversion of tryptophan to skatole and cyanide expands the mechanistic competence of diiron oxidases. J. Am. Chem. Soc. 147, 6326–6331 (2025).
Makris, T. M. et al. An unusual peroxo intermediate of the arylamine oxygenase of the chloramphenicol biosynthetic pathway. J. Am. Chem. Soc. 137, 1608–1617 (2015).
Rokob, T. A. Pathways for arene oxidation in non-heme diiron enzymes: lessons from computational studies on benzoyl coenzyme A epoxidase. J. Am. Chem. Soc. 138, 14623–14638 (2016).
Park, K. et al. Peroxide activation for electrophilic reactivity by the binuclear non-heme iron enzyme AurF. J. Am. Chem. Soc. 139, 7062–7070 (2017).
Wang, C. & Chen, H. Convergent theoretical prediction of reactive oxidant structures in diiron arylamine oxygenases AurF and CmlI: peroxo or hydroperoxo? J. Am. Chem. Soc. 139, 13038–13046 (2017).
Chalupský, J. et al. Reactivity of the binuclear non-heme iron active site of Δ9 desaturase studied by large-scale multireference ab initio calculations. J. Am. Chem. Soc. 136, 15977–15991 (2014).
Bím, D. et al. Proton-electron transfer to the active site is essential for the reaction mechanism of soluble Δ9-desaturase. J. Am. Chem. Soc. 142, 10412–10423 (2020).
Jasniewski, A. J. & Que, L. Dioxygen activation by nonheme diiron enzymes: diverse dioxygen adducts, high-valent intermediates, and related model complexes. Chem. Rev. 118, 2554–2592 (2018).
Fang, W. et al. Understanding the key roles of pH buffer in accelerating lignin degradation by lignin peroxidase. JACS Au 3, 536–549 (2023).
Poulos, T. L. Heme enzyme structure and function. Chem. Rev. 114, 3919–3962 (2014).
Pelletier, H. & Kraut, J. Crystal structure of a complex between electron transfer partners, cytochrome c peroxidase and cytochrome c. Science 258, 1748–1755 (1992).
Guengerich, F. P. Mechanisms of cytochrome P450-catalyzed oxidations. ACS Catal. 8, 10964–10976 (2018).
Bai, Y. et al. X-ray structure of a mammalian stearoyl-CoA desaturase. Nature 524, 252–256 (2015).
Chang, W. C. et al. Mechanism of the C5 stereoinversion reaction in the biosynthesis of carbapenem antibiotics. Science 343, 1140–1144 (2014).
Nakamura, H., Matsuda, Y. & Abe, I. Unique chemistry of non-heme iron enzymes in fungal biosynthetic pathways. Nat. Prod. Rep. 35, 633–645 (2018).
Marchand, J. A. et al. Discovery of a pathway for terminal-alkyne amino acid biosynthesis. Nature 567, 420–424 (2019).
Hedges, J. B. & Ryan, K. S. In vitro reconstitution of the biosynthetic pathway to the nitroimidazole antibiotic azomycin. Angew. Chem. Int. Ed. 58, 11647–11651 (2019).
Shimo, S. et al. Stereodivergent nitrocyclopropane formation during biosynthesis of belactosins and hormaomycins. J. Am. Chem. Soc. 143, 18413–18418 (2021).
Li, X., Shimaya, R., Dairi, T., Chang, W. C. & Ogasawara, Y. Identification of cyclopropane formation in the biosyntheses of hormaomycins and belactosins: sequential nitration and cyclopropanation by metalloenzymes. Angew. Chem. Int. Ed. 61, e202113189 (2022).
Simke, W. C. et al. Structural basis for methine excision by a heme oxygenase-like enzyme. ACS Cent. Sci. 10, 1524–1536 (2024).
Pope, S. R. et al. Heme oxygenase–like metalloenzymes. Annu. Rev. Biochem. 94, 59–88 (2025).
Kabsch, W. XDS. Acta. Crystallogr. D Struct. Biol. 66, 125–132 (2010).
Agirre, J. et al. The CCP4 suite: integrative software for macromolecular crystallography. Acta. Crystallogr. D Struct. Biol. 79, 449–461 (2023).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta. Crystallogr. D Struct. Biol. 66, 486–501 (2010).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta. Crystallogr. D Struct. Biol. 75, 861–877 (2019).
Frisch, M. J. et al. Gaussian 16 revision C.01 (Gaussian, 2016).
Marenich, A. V., Cramer, C. J. & Truhlar, D. G. Universal solvation model based on solute electron density and on a continuum model of the solvent defined by the bulk dielectric constant and atomic surface tensions. J. Phys. Chem. 113, 6378–6396 (2009).
Søndergaard, C. R., Olsson, M. H. M., Rostkowski, M. & Jensen, J. H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 7, 2284–2295 (2011).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004).
Bayly, C. I., Cieplak, P., Cornell, W. D. & Kollman, P. A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 97, 10269–10280 (1993).
Li, P. & Merz, K. M. J. MCPB.py: a Python based metal center parameter builder. J. Chem. Inf. Model. 56, 599–604 (2016).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3693–3713 (2015).
Kräutler, V., Van Gunsteren, W. F. & Hünenberger, P. H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 22, 501–508 (2001).
Case, D. A. et al. Amber 2018 (University of California, San Francisco, 2018).
Sherwood, P. et al. QUASI: a general purpose implementation of the QM/MM approach and its application to problems in catalysis. J. Mol. Struc. Theochem 632, 1–28 (2003).
Reinhart, A., Michael, B., Marco, H., Hans, H. & Christoph, K. Electronic structure calculations on workstation computers: the program system turbomole. Chem. Phys. Lett. 162, 165–169 (1989).
Smith, W. & Forester, T. R. DL_POLY_2.0: a general-purpose parallel molecular dynamics simulation package. J. Mol. Graph. 14, 136–141 (1996).
Bakowies, D. & Thiel, W. Hybrid models for combined quantum mechanical and molecular mechanical approaches. J. Phys. Chem. 100, 10580–10594 (1996).
Senn, H. M. & Thiel, W. QM/MM methods for biomolecular systems. Angew. Chem. Int. Ed. 48, 1198–1229 (2009).
Johannes Kästner et al. DL-FIND: an open-source geometry optimizer for atomistic simulations. J. Phys. Chem. A 113, 11856–11865 (2009).
Grimme, S. Accurate description of van der Waals complexes by density functional theory including empirical corrections. J. Comput. Chem. 25, 1463–1473 (2004).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (32470047 and 32170026 to W.C., 22122305 to B.W., 32371303 to Z.Z., 22403016 to J.L. and 22303043 to X. Zhang), the Scientific Research Innovation Capability Support Project for Young Faculty (ZYGXONJSKYCXNLZCXM-B6 to B.W.), the Hubei Province’s Outstanding Medical Academic Leader Program (Chutian Talent Plan) to W.C. and the China Postdoctoral Science Foundation (2023M742703 and GZC20231989 to R.G.). We are sincerely grateful to the staff members at the BL19U1, BL02U1 and BL10U2 beamlines of the National Facility for Protein Science in Shanghai (NFPS) for providing technical support in the X-ray diffraction data collection and analysis. We also acknowledge the staff members of the Electron Spin Resonance System at the Steady High Magnetic Field Facility for providing technical support and assistance with data collection, and the Core Facility of Wuhan University for LC-HRMS and NMR data collection.
Author information
Authors and Affiliations
Contributions
W.C., B.W. and Z.Z. conceived the project and directed the research. R.G. performed the genetic and biochemical experiments. Y.Q. conducted the structural biology studies. J.L., X. Zhang and Z.L. undertook the QM/MM analysis of the mechanism. L.Z., Z.T. and K.S. carried out the compound synthesis. X. Zeng and B.J. performed the purification of the compounds. L. Yu and R.C. assisted in preliminary genetic experiments. Y.Z. and L.L. carried out the stopped-flow experiments. Y.-S.C. analysed the NMR data. W.C., B.W., Z.Z., R.G., Y.Q. and J.L. wrote the manuscript; Z.D., X.S. and L. Yang made a critical reading of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemistry thanks Jennifer DuBois and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 [13C6]l-isoleucine-feeding analysis of E. coli BL21(DE3) recombinants.
a, Schematic of [13C6]l-isoleucine-feeding of the BL21(DE3)/polE + polF recombinant. b, LC-HRMS analysis of the target metabolite 1 produced by the BL21(DE3)/polE + polF recombinant fed with [13C6]l-isoleucine. The expected [M + H]+ ion for 1 and [13C6]-1 is at 128.0706 and 134.0907, respectively, whereas the detected [M + H]+ ion for 1 and [13C6]-1 is individually at 128.0711 and 134.0910. c, Schematic of [13C6]l-isoleucine-feeding of the BL21(DE3)/polF recombinant. d, LC-HRMS analysis of the target metabolite 1 produced by the BL21(DE3)/polF recombinant fed with [13C6]l-isoleucine. The expected [M + H]+ ion for 1 and [13C6]-1 is at 128.0706 and 134.0907, respectively, whereas the detected [M + H]+ ion for 1 and [13C6]-1 is individually at 128.0706 and 134.0907. e, Schematic of [13C6]l-isoleucine-feeding of the BL21(DE3)/polE recombinant. f, LC-HRMS analysis of the target metabolite 2 produced by the BL21(DE3)/polE recombinant fed with [13C6]l-isoleucine. The expected [M + H]+ ion for 2 and [13C6]-2 is at 130.0863 and 136.1064, respectively, whereas the detected [M + H]+ ion for 2 and [13C6]-2 is correspondingly at 130.0866 and 136.1067.
Extended Data Fig. 2 In vitro characterization of PolE as an unusual Fe2+/pterin-dependent desaturase.
a, Schematic of the reaction catalyzed by PolE. b, PolE reactions with individual addition of BH4, 6-MePH4, and THF. Std, the authentic l-isoleucine and 2 standards; +BH4, the reaction with Fe2+, BH4, and FolA added; +6-MePH4, the reaction with Fe2+, 6-MePH4, and FolA added; +THF, the reaction with Fe2+, THF, and FolA added; NC, the reaction without PolE added as negative control. c, Evaluation of the metal dependence of PolE. PolE was firstly chelated with EDTA, and then its reactions were individually supplemented with various divalent metal ions. d, Schematic of the hypothetic catalytic-process by PolE. e, LC-HRMS analysis of the PolE reactions using 3 or 4 as substrate. Std, the authentic 2, 3 and 4 standards; +(3+PolE), the PolE reaction using 3 as substrate; +3, the reaction without PolE added as negative control; +(4+PolE), the PolE reaction using 4 as substrate; +4, the reaction without PolE added as negative control. f, LC-HRMS analysis of the PolE reactions using 5 or 6 as substrate. Std, the authentic 2, 5 and 6 standards; +(5+PolE), the PolE reaction using 5 as substrate; +5, the reaction without PolE added as negative control; +(6+PolE), the PolE reaction using 6 as substrate; +6, the reaction without PolE added as negative control.
Extended Data Fig. 3 Structural characterization of SnPolE and functional analysis of its substrate-coordination sites.
a, Surface representation of SnPolE with Fe (orange sphere), BH4 (purple stick), and l-isoleucine (blue stick). b, The structure alignment of apo SnPolE (pink cartoon), SnPolE-BH4 (yellow cartoon), and SnPolE-BH4-l-isoleucine (green cartoon). c, Electron map of apo SnPolE. The 2Fo − Fc map is shown for Fe-coordinating residues and water (grey mesh, contoured at 1.0σ). Fe-coordinating residues are shown in lines and sticks, Fe is illustrated as orange sphere, and water is indicated as red sphere. d, Electron map of ligands bond in SnPolE-BH4 binary complex. The 2Fo − Fc map is shown for Fe-coordinating residues, water, and BH4 (grey mesh, contoured at 1.0σ). BH4 is shown in purple stick. e, Electron map of ligands bound in the SnPolE-BH4-l-isoleucine ternary complex structure. The Fo − Fc omit maps of l-isoleucine (cyan stick), Fe ions (orange spheres), the Fe-coordinating water molecule (red sphere) and Fe-coordinating residues (sticks) are contoured at 2.5σ (green mesh). f, Comparison of the binding pockets of apo SnPolE, SnPolE-BH4 binary complex, and SnPolE-BH4-l-isoleucine ternary complex. SnPolE, SnPolE-BH4, SnPolE-BH4-l-isoleucine is correspondingly shown in pink, yellow, and green. g, Crystal structure of SnPolEY157A variant with l-isoleucine mistakenly coordinated to iron. The selected amino acid residues and l-isoleucine are shown in the stick and ball, and Fe is illustrated as sphere. h, Comparison of active sites of SnPolEY157A (yellow) and SnPolE (white). i, Comparison of active sites of SnPolEF140A (deep teal) and SnPolE (white). j, Comparison of active sites of SnPolEY280A (pink) and SnPolE (white).
Extended Data Fig. 4 Mechanistic investigations of SnPolE.
a, The potential energy surface (in kcal mol−1) of SnPolE-catalyzed desaturation reaction obtained from QM/MM calculations. b, The structures of key intermediates involved in the SnPolE-catalyzed desaturation reaction.
Extended Data Fig. 5 In vitro characterization and monitoring of the catalytic process of PolF.
a, Schematic of the proposed catalytic process by PolF. b, LC-HRMS analysis of PolF activity in the presence of Fe2+ or other divalent metal ions. PolF was firstly chelated with EDTA, and then its reactions were individually supplemented with various divalent metal ions. c, LC-HRMS analysis of PolF reactions with l-isoleucine as substrate. Activity assays of PolF were performed under aerobic atmosphere. Std, the authentic standards of l-isoleucine and 1; Full reaction, the PolF reaction with Fe2+ and VC added; -O2, the full reaction incubated under anaerobic atmosphere; -Fe2+, the PolF reaction without Fe2+ added; -Fe2+(+EDTA), the PolF reaction with EDTA added; -VC, reaction without VC added; -PolF, the reaction without PolF added; -l-Ile, the reaction without l-isoleucine added. d, LC-HRMS analysis of the PolF reactions using 3 and 4 as substrate. Std, the authentic standards of 3, 4, and 1; +(3+PolF), the PolF reaction using 3 as substrate; +3, the reaction without PolF added as negative control; +(4+PolF), the PolF reaction using 4 as substrate; +4, the reaction without PolF added as negative control. e, LC-HRMS analysis of the PolF reactions using 5 and 6 as substrate. Std, the authentic standards of 5, 6, and 1; +(5+PolF), the PolF reaction using 5 as substrate; +5, the reaction without PolF added as negative control; +(6+PolF), the PolF reaction using 6 as substrate; +6, the reaction without PolF added as negative control.
Extended Data Fig. 6 Structure comparison of PolF and its homologues.
a, Overall structure of Fe2(II/II)-PolF (yellow). b, Overall structure of Fe(II)-SaPolF (blue). c, Overall structure of Fe2(II/II)-SzPolF (green). d, Structure alignment of PolF, SaPolF, and SzPolF with highlight of the core helix α3.
Extended Data Fig. 7 Comparison of the active pocket sites of Fe(II)-SzPolF and Fe2(II/II)-SzPolF.
a, The three core α helices (core α1, α2, and α3) harbor metal-binding ligands. b, Fe coordination sites of Fe2(II/II)-SzPolF. Six-coordinate Fe1 has an overall octahedral geometry, whereas the five-coordinate Fe2 attains roughly square pyramidal symmetry. c, Comparison of the binding pockets of Fe(II)-SzPolF and Fe2(II/II)-SzPolF. Fe(II)-SzPolF is shown in purple sticks and Fe2(II/II)-SzPolF is shown in yellow sticks. d, Slice-surface view of Fe(II)-SzPolF (left) and Fe2(II/II)-SzPolF (right). Electrostatic surface potential is colored by red (-79.400 kT/e), blue (+79.400 kT/e), and white (neutral). e, Electron map of ligand bond in Fe(II)-SzPolF-2 complex. Structure of Fe(II)-SzPolF reveals that Fe1 (orange sphere) recruits 2 (green sticks) as a bidentate ligand. Selected amino acids are shown in green stick format. The 2Fo − Fc map is shown for residues, Fe1 and substrate (grey mesh, contoured at 1.0σ). f, Hydrogen-bonding and hydrophobic-interaction with selected amino acids and 2.
Extended Data Fig. 8 Stopped-flow absorption spectroscopy demonstrates the accumulation of intermediate in the SzPolF reaction.
Absorption spectra acquired after rapid mixing an O2-free solution of SzPolF (0.3 mM) and Fe2+ (0.6 mM, 2 molar equiv.) in the absence (a) or presence of (b) 2 mM 2 (without VC), (c) 2 mM 2 (with VC), (d) 2 mM l-isoleucine (with VC), with an equal volume of O2-saturated buffer.
Extended Data Fig. 9 Mechanistic investigations of SzPolF catalyzed desaturation in the presence of reductants.
a, The potential energy surface (in kcal mol−1) of SzPolF-catalyzed desaturation reaction obtained from QM/MM calculations. b, The structures of key intermediates involved in the SzPolF-catalyzed desaturation reaction.
Extended Data Fig. 10 Distribution of selected PolF homologues in microbial genomes.
a, Phylogenetic analysis of PolF and its homologues with diverse microbial origins. The branches corresponding to PolF, SaPolF, SrPolF, and SzPolF are highlighted. b, Selected biosynthetic gene clusters that encode the homologues of PolF.
Supplementary information
Supplementary Information
Supplementary Figs. 1–132, Tables 1–15 and Methods.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gong, R., Qu, Y., Liu, J. et al. A two-metalloenzyme cascade constructs the azetidine-containing pharmacophore. Nat. Chem. (2025). https://doi.org/10.1038/s41557-025-01949-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41557-025-01949-y
This article is cited by
-
Structural and mechanistic insights into azetidine-associated αKG-NHFe enzyme OkaE with multifunctional catalysis
Nature Communications (2026)