Abstract
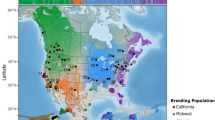
Differences in life history can cause co-distributed species to evolve contrasting population genetic patterns, even as they occupy the same landscape. In high-latitude animals, evolutionary processes may be especially influenced by long-distance seasonal migration, a widespread adaptation to seasonality. Although migratory movements are intuitively linked to dispersal and therefore promotion of gene flow, their evolutionary genetic consequences remain poorly understood. Using ~1,700 genomes from 35 co-distributed boreal-breeding bird species that differ in non-breeding latitude and thus migration distance, we find that most long-distance migrants unexpectedly exhibit spatial genetic structure, despite their strong movement propensity. This result suggests evolutionary effects of philopatry—the tendency of many migrants to return to the same breeding site year after year, resulting in restricted dispersal. We further demonstrate that migration distance and genetic diversity are strongly positively correlated in our study species. This striking relationship suggests that the adaptive seasonal shifts in biogeography inherent to long-distance migration may enhance population stability, preserving genetic diversity in long-distance migrants relative to shorter-distance migrants that winter in harsher conditions at higher latitudes. Our results suggest that the major impact of long-distance seasonal migration on population genetic evolution occurs through promotion of demographic stability, rather than facilitation of dispersal.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The sequence data generated during the study have been uploaded to the NCBI Sequence Read Archive under BioProjects PRJNA1043688 and PRJNA1130443. Individual accession numbers for each sample are provided in Supplementary Dataset 1. All other data are available within the manuscript, the repositories described in the Code availability statement and the Supplementary Information files. Source data are provided with this paper.
Code availability
Bioinformatic code used to process sequence data are available via figshare at https://doi.org/10.6084/m9.figshare.27284553 (ref. 119). Code used to conduct comparative analyses are available via a Code Ocean capsule at https://doi.org/10.24433/CO.5578409.v1 (ref. 120).
References
Avise, J. C. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 18, 489–522 (1987).
Papadopoulou, A. & Knowles, L. L. Toward a paradigm shift in comparative phylogeography driven by trait-based hypotheses. Proc. Natl. Acad. Sci. USA 113, 8018–8024 (2016).
Romiguier, J. et al. Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature 515, 261–263 (2014).
Bradburd, G. S. & Ralph, P. L. Spatial population genetics: it’s about time. Annu. Rev. Ecol. Evol. Syst. 50, 427–449 (2019).
Bachmann, J. C., Jansen van Rensburg, A., Cortazar-Chinarro, M., Laurila, A. & van Buskirk, J. Gene flow limits adaptation along steep environmental gradients. Am. Nat. 195, E67–E86 (2020).
Singhal, S. et al. Does population structure predict rate of speciation? A comparative test across Australia’s most diverse vertebrate radiation. Am. Nat. 192, 432–447 (2018).
Edwards, S. V., Robin, V. V., Ferrand, N. & Moritz, C. The evolution of comparative phylogeography: putting the geography (and more) into comparative population genomics. Genome Biol. Evol. 14, evab176 (2022).
Gagnaire, P. A. Comparative genomics approach to evolutionary process connectivity. Evol. Appl. 13, 1320–1334 (2020).
Zbinden, Z. D., Douglas, M. R., Chafin, T. K. & Douglas, M. E. Riverscape community genomics: a comparative analytical approach to identify common drivers of spatial structure. Mol. Ecol. 32, 6743–6765 (2022).
Riginos, C. & Jahnke, M. Comparative landscape genomics has arrived with a splash. Mol. Ecol. 32, 6725–6728 (2023).
Ralston, J. & Kirchman, J. J. Continent-scale genetic structure in a boreal forest migrant, the blackpoll warbler (Setophaga striata). Auk 129, 467–478 (2012).
Weir, J. T. & Schluter, D. Ice sheets promote speciation in boreal birds. Proc. R. Soc. B 271, 1881–1887 (2004).
Ruegg, K. C. & Smith, T. B. Not as the crow flies: a historical explanation for circuitous migration in Swainson’s thrush (Catharus ustulatus). Proc. R. Soc. B 269, 1375–1381 (2002).
Graham, B. A. & Burg, T. M. Molecular markers provide insights into contemporary and historic gene flow for a non-migratory species. J. Avian Biol. 43, 198–214 (2012).
Ralston, J. et al. Comparative phylogeographic analysis suggests a shared history among eastern North American boreal forest birds. Ornithology 138, ukab018 (2021).
Slatkin, M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47, 264–279 (1993).
Winger, B. M. & Pegan, T. M. Migration distance is a fundamental axis of the slow-fast continuum of life history in boreal birds. Ornithology 138, ukab043 (2021).
Fonseca, E. M., Pelletier, T. A., Decker, S. K., Parsons, D. J. & Carstens, B. C. Pleistocene glaciations caused the latitudinal gradient of within-species genetic diversity. Evol. Lett. 7, 331–338 (2023).
Paradis, E., Baillie, S. R., Sutherland, W. J. & Gregory, R. D. Patterns of natal and breeding dispersal in birds. J. Anim. Ecol. 67, 518–536 (1998).
Weeks, B. C. et al. Morphological adaptations linked to flight efficiency and aerial lifestyle determine natal dispersal distance in birds. Funct. Ecol. 36, 1681–1689 (2022).
Turbek, S. P., Scordato, E. S. C. & Safran, R. J. The role of seasonal migration in population divergence and reproductive isolation. Trends Ecol. Evol. 33, 164–175 (2018).
Winker, K. On the origin of species through heteropatric differentiation: a review and a model of speciation in migratory animals. Ornithol. Monogr. 69, 1–30 (2010).
Chu, J. J. & Claramunt, S. Determinants of natal dispersal distances in North American birds. Ecol. Evol. 13, e9789 (2023).
Toews, D. P. L. Habitat suitability and the constraints of migration in New World warblers. J. Avian Biol. 48, 1614–1623 (2017).
Bensch, S. Is the range size of migratory birds constrained by their migratory program? J. Biogeogr. 26, 1225–1235 (1999).
Greenberg, R. & Marra, P. P. (eds) Birds of Two Worlds: The Ecology and Evolution of Migration (Johns Hopkins Univ. Press, 2005).
Kimmitt, A. A., Pegan, T. M., Jones, A. W., Winker, K. & Winger, B. M. How veeries vary: whole genome sequencing resolves genetic structure in a long-distance migratory bird. Ornithology 141, ukad061 (2023).
Johnson, O. et al. Amazonian birds in more dynamic habitats have less population genetic structure and higher gene flow. Mol. Ecol. 32, 2186–2205 (2023).
Wilkins, J. F. A separation-of-timescales approach to the coalescent in a continuous population. Genetics 168, 2227–2244 (2004).
Hewitt, G. M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 58, 247–276 (1996).
Dyke, A. S. & Prest, V. K. Late Wisconsinan and Holocene history of the Laurentide ice sheet. Géogr. Phys. Quatern. 41, 237–263 (1987).
Nei, M., Maruyama, T. & Chakraborty, R. The bottleneck effect and genetic variability in populations. Evolution 29, 1–10 (1975).
Frankham, R. Genetics and extinction. Biol. Conserv. 126, 131–140 (2005).
García-Berro, A. et al. Migratory behaviour is positively associated with genetic diversity in butterflies. Mol. Ecol. 32, 560–574 (2023).
Pegan, T. M., Berv, J. S., Gulson-Castillo, E. R., Kimmitt, A. A. & Winger, B. M. The pace of mitochondrial molecular evolution varies with seasonal migration distance. Evolution 78, 160–173 (2024).
Ellegren, H. & Galtier, N. Determinants of genetic diversity. Nat. Rev. Genet. 17, 422–433 (2016).
Mackintosh, A. et al. The determinants of genetic diversity in butterflies. Nat. Commun. 10, 3466 (2019).
Kimmitt, A. A. et al. Genetic evidence for widespread population size expansion in North American boreal birds prior to the Last Glacial Maximum. Proc. R. Soc. B 290, 20221334 (2023).
Ovaskainen, O. et al. How to make more out of community data? A conceptual framework and its implementation as models and software. Ecol. Lett. 20, 561–576 (2017).
Novembre, J. et al. Genes mirror geography within Europe. Nature 456, 98–102 (2008).
Winger, B. M., Auteri, G. G., Pegan, T. M. & Weeks, B. C. A long winter for the Red Queen: rethinking the evolution of seasonal migration. Biol. Rev. 94, 737–752 (2019).
Kokko, H. Competition for early arrival in migratory birds. J. Anim. Ecol. 68, 940–950 (1999).
Stamps, J. A. & Davis, J. M. Adaptive effects of natal experience on habitat selection by dispersers. Anim. Behav. 72, 1279–1289 (2006).
McNamara, J. M. & Dall, S. R. X. The evolution of unconditional strategies via the ‘multiplier effect’. Ecol. Lett. 14, 237–243 (2011).
DeLuca, W. V. et al. A boreal songbird’s 20,000 km migration across North America and the Atlantic Ocean. Ecology 100, e02651 (2019).
Conklin, J. R. et al. High dispersal ability versus migratory traditions: fine-scale population structure and post-glacial colonisation in bar-tailed godwits. Mol. Ecol. 33, e17452 (2024).
Luna, L. W. et al. Late Pleistocene landscape changes and habitat specialization as promoters of population genomic divergence in Amazonian floodplain birds. Mol. Ecol. 32, 214–228 (2023).
Talavera, A. & Tellería, J. L. Does microhabitat use affect population differentiation? A test with southwestern Palaearctic forest birds. J. Ornithol. 163, 923–929 (2022).
Salisbury, C. L., Seddon, N., Cooney, C. R. & Tobias, J. A. The latitudinal gradient in dispersal constraints: ecological specialisation drives diversification in tropical birds. Ecol. Lett. 15, 847–855 (2012).
Ovaskainen, O. & Abrego, N. Joint Species Distribution Modeling With Applications in R (Cambridge Univ. Press, 2020).
Felsenstein, J. Phylogenies and the comparative method. Am. Nat. 125, 1–15 (1985).
Medina, I., Cooke, G. M. & Ord, T. J. Walk, swim or fly? Locomotor mode predicts genetic differentiation in vertebrates. Ecol. Lett. 21, 638–645 (2018).
D’Urban Jackson, J. et al. Polygamy slows down population divergence in shorebirds. Evolution 71, 1313–1326 (2017).
Kwon, E. et al. Breeding site fidelity is lower in polygamous shorebirds and male-biased in monogamous species. Behav. Ecol. 33, 592–605 (2022).
Li, M. H. & Merilä, J. Genetic evidence for male-biased dispersal in the Siberian jay (Perisoreus infaustus) based on autosomal and Z-chromosomal markers. Mol. Ecol. 19, 5281–5295 (2010).
Drever, M. C., Smith, A. C., Venier, L. A., Sleep, D. J. H. & MacLean, D. A. Cross-scale effects of spruce budworm outbreaks on boreal warblers in eastern Canada. Ecol. Evol. 8, 7334–7345 (2018).
Pierson, J. C., Allendorf, F. W., Drapeau, P. & Schwartz, M. K. Breed locally, disperse globally: fine-scale genetic structure despite landscape-scale panmixia in a fire-specialist. PLoS ONE 8, e67248 (2013).
Orme, D. et al. Caper: comparative analyses of phylogenetics and evolution in R https://cran.r-project.org/web/packages/caper/index.html (2012).
Korunes, K. L. & Samuk, K. pixy: unbiased estimation of nucleotide diversity and divergence in the presence of missing data. Mol. Ecol. Resour. 21, 1359–1368 (2021).
Prasad, A., Lorenzen, E. D. & Westbury, M. V. Evaluating the role of reference-genome phylogenetic distance on evolutionary inference. Mol. Ecol. Resour. 22, 45–55 (2022).
Thorburn, D. M. J. et al. Origin matters: using a local reference genome improves measures in population genomics. Mol. Ecol. Resour. 23, 1706–1723 (2023).
Vurture, G. W. et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33, 2202–2204 (2017).
Excoffier, L. in Handbook of Statistical Genetics 3rd edn (eds Balding, D. J. et al.) 980–1020 (Wiley, 2007).
Pannell, J. R. & Charlesworth, B. Effects of metapopulation processes on measures of genetic diversity. Phil. Trans. R. Soc. B 355, 1851–1864 (2000).
Banks, S. C. et al. How does ecological disturbance influence genetic diversity? Trends Ecol. Evol. 28, 670–679 (2013).
Calvert, A. M., Walde, S. J. & Taylor, P. D. Nonbreeding-season drivers of population dynamics in seasonal migrants: conservation parallels across taxa. Avian Conserv. Ecol. 4, 5 (2009).
Forsman, J. T. & Mönkkönen, M. The role of climate in limiting European resident bird populations. J. Biogeogr. 30, 55–70 (2003).
Both, C. et al. Avian population consequences of climate change are most severe for long-distance migrants in seasonal habitats. Proc. R. Soc. B 277, 1259–1266 (2010).
De Meester, L., Stoks, R. & Brans, K. I. Genetic adaptation as a biological buffer against climate change: potential and limitations. Integr. Zool. 13, 372–391 (2018).
Omernik, J. M. & Griffith, G. E. Ecoregions of the conterminous United States: evolution of a hierarchical spatial framework. Environ. Manag. 54, 1249–1266 (2014).
Cumming, S. G. et al. Climate and vegetation hierarchically structure patterns of songbird distribution in the Canadian boreal region. Ecography 37, 137–151 (2014).
Stralberg, D. et al. Biogeography of boreal passerine range dynamics in western North America: past, present, and future. Ecography 40, 1050–1066 (2017).
Fair, J., Paul, E., Jones, J. & Bies, L. (eds) Guidelines to the Use of Wild Birds in Research (Ornithological Council, 2023).
Milá, B., Smith, T. B. & Wayne, R. K. Speciation and rapid phenotypic differentiation in the yellow-rumped warbler Dendroica coronata complex. Mol. Ecol. 16, 159–173 (2007).
Milá, B., McCormack, J. E., Castañeda, G., Wayne, R. K. & Smith, T. B. Recent postglacial range expansion drives the rapid diversification of a songbird lineage in the genus Junco. Proc. R. Soc. B 274, 2653–2660 (2007).
Colbeck, G. J., Gibbs, H. L., Marra, P. P., Hobson, K. & Webster, M. S. Phylogeography of a widespread North American migratory songbird (Setophaga ruticilla). J. Hered. 99, 453–463 (2008).
van Els, P., Spellman, G. M., Smith, B. T. & Klicka, J. Extensive gene flow characterizes the phylogeography of a North American migrant bird: black-headed grosbeak (Pheucticus melanocephalus). Mol. Phylogenet. Evol. 78, 148–159 (2014).
Hindley, J. A., Graham, B. A., Pulgarin-R, P. C. & Burg, T. M. The influence of latitude, geographic distance, and habitat discontinuities on genetic variation in a high latitude montane species. Sci. Rep. 8, 11846 (2018).
Miller, C. V. et al. Genomics-informed conservation units reveal spatial variation in climate vulnerability in a migratory bird. Mol. Ecol. 33, e17199 (2023).
Perez, M. F. et al. Assessing population structure in the face of isolation by distance: are we neglecting the problem? Divers. Distrib. 24, 1883–1889 (2018).
Schweizer, T. M. & DeSaix, M. G. Cost-effective library preparation for whole genome sequencing with feather DNA. Conserv. Genet. Resour. 15, 21–28 (2023).
Therkildsen, N. O. & Palumbi, S. R. Practical low-coverage genomewide sequencing of hundreds of individually barcoded samples for population and evolutionary genomics in nonmodel species. Mol. Ecol. Resour. 17, 194–208 (2017).
Schubert, M., Lindgreen, S. & Orlando, L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes 9, 88 (2016).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Lou, R. N. & Therkildsen, N. O. Batch effects in population genomic studies with low-coverage whole genome sequencing data: causes, detection and mitigation. Mol. Ecol. Resour. 22, 1678–1692 (2021).
Zhang, G. et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346, 1311–1321 (2014).
Ruegg, K. et al. Ecological genomics predicts climate vulnerability in an endangered southwestern songbird. Ecol. Lett. 21, 1085–1096 (2018).
Toews, D. P. L. et al. Plumage genes and little else distinguish the genomes of hybridizing warblers. Curr. Biol. 26, 2313–2318 (2016).
Feng, S. et al. Dense sampling of bird diversity increases power of comparative genomics. Nature 587, 252–257 (2020).
Laine, V. N. et al. Evolutionary signals of selection on cognition from the great tit genome and methylome. Nat. Commun. 7, 10474 (2016).
Manthey, J. D., Klicka, J. & Spellman, G. M. The genomic signature of allopatric speciation in a songbird is shaped by genome architecture (Aves: Certhia americana). Genome Biol. Evol. 13, evab120 (2021).
Friis, G., Vizueta, J., Ketterson, E. D. & Milá, B. A high-quality genome assembly and annotation of the dark-eyed junco Junco hyemalis, a recently diversified songbird. G3 12, jkac083 (2022).
Sly, N. D. et al. Molecular parallelism in signaling function across different sexually selected ornaments in a warbler. Proc. Natl Acad. Sci. USA 119, e2120482119 (2022).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Jun, G., Wing, M. K., Abecasis, G. & Kang, H. M. An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res. https://doi.org/10.1101/gr.176552.114 (2015).
Van der Auwera, G. A. et al. From fastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. https://doi.org/10.1002/0471250953.bi1110s43 (2013).
Dierckxsens, N., Mardulyn, P. & Smits, G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18 (2016).
Korneliussen, T. S., Albrechtsen, A. & Nielsen, R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinform. 15, 356 (2014).
Linderoth, T. Identifying Population Histories, Adaptive Genes, and Genetic Duplication from Population-Scale Next Generation Sequencing. PhD dissertation, Univ. California, Berkeley (2018).
Meisner, J. & Albrechtsen, A. Inferring population structure and admixture proportions in low-depth NGS data. Genetics 210, 719–731 (2018).
Harringmeyer, O. S. & Hoekstra, H. E. Chromosomal inversion polymorphisms shape the genomic landscape of deer mice. Nat. Ecol. Evol. 6, 1965–1979 (2022).
Ishigohoka, J. et al. Distinct patterns of genetic variation at low-recombining genomic regions represent haplotype structure. Evolution 78, 1916–1935 (2024).
Li, H. & Ralph, P. Local PCA shows how the effect of population structure differs along the genome. Genetics 211, 289–304 (2019).
Privé, F., Luu, K., Blum, M. G. B., McGrath, J. J. & Vilhjálmsson, B. J. Efficient toolkit implementing best practices for principal component analysis of population genetic data. Bioinformatics 36, 4449–4457 (2020).
Waters, P. D. et al. Microchromosomes are building blocks of bird, reptile, and mammal chromosomes. Proc. Natl Acad. Sci. USA 118, e2112494118 (2021).
Hijmans, R. J. Package ‘geosphere’ https://cran.r-project.org/web/packages/geosphere/index.html (2022).
Pegan, T. M. & Winger, B. M. The influence of seasonal migration on range size in temperate North American passerines. Ecography 43, 1191–1202 (2020).
Dunning, J. B. J. CRC Handbook of Avian Body Masses (CRC Press, 2008).
Birds of the World (Cornell Lab of Ornithology, 2022).
Rasmussen, M. S., Garcia-Erill, G., Korneliussen, T. S., Wiuf, C. & Albrechtsen, A. Estimation of site frequency spectra from low-coverage sequencing data using stochastic EM reduces overfitting, runtime, and memory usage. Genetics 222, iyac148 (2022).
Korneliussen, T. S. Heterozygosity https://www.popgen.dk/angsd/index.php/Heterozygosity (2017).
Zimova, M. et al. Body size predicts the rate of contemporary morphological change in birds. Proc. Natl Acad. Sci. USA 120, e2206971120 (2023).
Jetz, W., Thomas, G. H., Joy, J. B., Hartmann, K. & Mooers, A. O. The global diversity of birds in space and time. Nature 491, 444–448 (2012).
Rubolini, D., Liker, A., Garamszegi, L. Z., Moller, A. P. & Saino, N. Using the BirdTree.org website to obtain robust phylogenies for avian comparative studies: a primer. Curr. Zool. 61, 959–965 (2015).
Youngflesh, C. MCMCvis: tools to visualize, manipulate, and summarize MCMC output. J. Open Source Softw. 3, 640 (2018).
Gabry, J., Simpson, D., Vehtari, A., Betancourt, M. & Gelman, A. Visualization in Bayesian workflow. J. R. Stat. Soc. A 182, 389–402 (2019).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Pegan, T. M. et al. Bioinformatic code for manuscript “Seasonal migration to the tropics promotes genetic diversity but not gene flow in boreal birds”. figshare https://doi.org/10.6084/m9.figshare.27284553 (2025).
Pegan, T. M. et al. Seasonal migration to the tropics promotes genetic diversity but not gene flow in boreal birds. Code Ocean https://doi.org/10.24433/CO.5578409.v1 (2025).
Acknowledgements
For comments and advice, we thank G. Bradburd, R. Burner, K. Wacker, E. Gulson-Castillo, V. Gómez-Bahamón, J. Berv, Z. Hancock, M. Hack, A. Marshall, L. Knowles, D. Rabosky, M. Witynksi and A. Benavides. For providing samples, we thank the curators, collections staff and field collectors from the following institutions: American Museum of Natural History, Cleveland Museum of Natural History, Cornell University Museum of Vertebrates, New York State Museum, Royal Alberta Museum, Royal Ontario Museum, University of Alaska Museum of the North, University of Minnesota Museum of Natural History, University of California, Berkeley Museum of Vertebrate Zoology, University of Michigan Museum of Zoology and University of Washington Burke Museum. For additional samples, we thank J. Tremblay (Environment and Climate Change Canada). For field permits, we thank the United States Fish and Wildlife Service, the United States Forest Service, the Minnesota Department of Natural Resources, the Michigan Department of Natural Resources, the Canadian Wildlife Service of Environment and Climate Change Canada, Alberta Fish and Wildlife, and Manitoba Fish and Wildlife. Field sampling was approved by the University of Michigan Animal Care and Use Committee (no. PRO00010608). For assistance in the field, we thank C. Brennan, S. Campbell, S. DuBay, G. M. Erickson, M. M. Ferraro, A. FitzGerald, L. Gooch, E. Gulson-Castillo, J. Ralston, C. Scobie, H. Skeen, V. Ting, K. Wacker and members of the Burg lab. For assistance in the lab, we thank T. Schweizer, C. Rayne, R. Herman, J. Yan, C. Kaczmarek, M. Florkowski, M. Guza, C. Pajka, M. Hack and C. Jordan. Next-generation sequencing for this project was partially carried out in the Advanced Genomics Core at the University of Michigan. This research was also supported in part through computational resources and services provided by Advanced Research Computing (ARC), a division of Information and Technology Services (ITS) at the University of Michigan, Ann Arbor. Funding includes National Science Foundation grant DEB 2146950 (B.M.W.), National Science Foundation Graduate Research Fellowship DGE 1256260, Fellow ID 2018240490 (T.M.P.), Jean Wright Cohn Endowment Fund at the University of Michigan Museum of Zoology, Robert W. Storer Endowment Fund at the University of Michigan Museum of Zoology, Mary Rhoda Swales Museum of Zoology Research Fund at the University of Michigan Museum of Zoology, William G. Fargo Fund at the University of Michigan Museum of Zoology, William A. and Nancy R. Klamm Endowment funds at the Cleveland Museum of Natural History, University of Michigan Rackham Graduate Student Research Grant (T.M.P.), British Ornithologists Union Small Research Grant (T.M.P.), and American Museum of Natural History Chapman Research Grant (T.M.P.).
Author information
Authors and Affiliations
Contributions
B.M.W. and T.M.P conceived the study. Genomic samples were contributed by all authors and were prepared for sequencing by T.M.P. and A.A.K. with methodological support from K.C.R. Data were analysed and visualized by T.M.P. and B.M.W. T.M.P. and B.M.W. wrote the paper with input and revisions from all authors. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Ecology & Evolution thanks Leilton Luna and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The phylogenetic relationship among the species in the study and species attributes included in HMSC models.
The phylogenetic relationship is shown on the tree from birdtree.org used in our analyses. The tree is plotted alongside a heatmap representing interspecific variation in the four species attributes we investigated in HMSC models (migration distance, mass, association with early successional habitat, and genetic diversity). Genetic diversity estimates were calculated in this study (Supplementary Table 1) and values for the other three attributes come from published sources as described in the Methods.
Extended Data Fig. 2 Migration distance correlates with an estimate of heterozygosity from higher-coverage samples.
Each point represents one species (N = 27) and points are colored by migration distance, as in main text figures, where non-migrants are shown in gray. This plot shows heterozygosity estimated by ANGSD from reference-mapped genomes. The intercept and slope of the line were estimated with a phylogenetic generalized least squares (PGLS) model.
Supplementary information
Supplementary Information
Legends for Supplementary Datasets 1–3, legends for Supplementary Figs. 1–35 and Supplementary Figs. 1–35.
Supplementary Data 1
List of samples used and metadata for each sample.
Supplementary Data 2
Bioinformatic metadata for each species.
Supplementary Data 3
Information about chromosomes and large genomic regions excluded from analyses based on evidence for possible inversion polymorphism.
Supplementary Data 4
Statistical Source Data for Supplementary Figs. 1–35 in three tabs. The first tab includes data underlying PCA plots and admixture plots (A and B panels), as well as lat/long data shown in panel C. The second tab includes data underlying the scatterplots (D panels). The third tab contains the slope and intercept values plotted on each scatterplot (D panels). Data for all figures are provided together and can be separated using the ‘Species’ column.
Source data
Source Data Fig. 3
Statistical Source Data. Excel file includes HMSC output data for each panel. The Excel file contains 12 tabs, with 4 tabs per panel, reflecting the 4 model chains we ran and then plotted using mcmc_intervals in the R package bayesplot.
Source Data Extended Data Fig. 1
Source file provides the phylogenetic tree. All data plotted alongside the tree is given in Extended_Data_Table1.pdf.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pegan, T.M., Kimmitt, A.A., Benz, B.W. et al. Long-distance seasonal migration to the tropics promotes genetic diversity but not gene flow in boreal birds. Nat Ecol Evol 9, 957–969 (2025). https://doi.org/10.1038/s41559-025-02699-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41559-025-02699-3