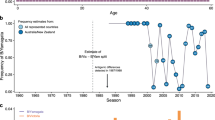
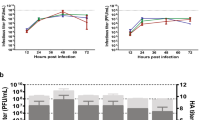
Abstract
Influenza exposures early in life are believed to shape future susceptibility to influenza infections by imprinting immunological biases that affect cross-reactivity to future influenza viruses. However, direct serological evidence linked to susceptibility is limited. Here we analysed haemagglutination-inhibition titres in 1,451 cross-sectional samples collected between 1992 and 2020, from individuals born between 1917 and 2008, against influenza B virus (IBV) isolates from 1940 to 2021. We included testing of ‘future’ isolates that circulated after sample collection. We show that immunological biases are conferred by early life IBV infection and result in lineage-specific cross-reactivity of a birth cohort towards future IBV isolates. This translates into differential estimates of susceptibility between birth cohorts towards the B/Yamagata and B/Victoria lineages, predicting lineage-specific birth-cohort distributions of observed medically attended IBV infections. Our data suggest that immunological measurements of imprinting could be important in modelling and predicting virus epidemiology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The raw HI datasets used for all analyses and figures are available via Zenodo at https://doi.org/10.5281/zenodo.10633085 (ref. 53). Source data are provided with this paper.
Code availability
The R codes used for all analyses and figures are available via Zenodo at https://doi.org/10.5281/zenodo.10633085 (ref. 53).
References
Cobey, S. & Hensley, S. E. Immune history and influenza virus susceptibility. Curr. Opin. Virol. 22, 105–111 (2017).
Oidtman, R. J. et al. Influenza immune escape under heterogeneous host immune histories. Trends Microbiol. 29, 1072–1082 (2021).
Gostic, K. M., Ambrose, M., Worobey, M. & Lloyd-Smith, J. O. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 354, 722–726 (2016).
Worobey, M., Han, G. Z. & Rambaut, A. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc. Natl Acad. Sci. USA 111, 8107–8112 (2014).
Gostic, K. M. et al. Childhood immune imprinting to influenza A shapes birth year-specific risk during seasonal H1N1 and H3N2 epidemics. PLoS Pathog. 15, e1008109 (2019).
Arevalo, P., McLean, H. Q., Belongia, E. A. & Cobey, S. Earliest infections predict the age distribution of seasonal influenza A cases. eLife 9, e50060 (2020).
Vieira, M. C. et al. Lineage-specific protection and immune imprinting shape the age distributions of influenza B cases. Nat. Commun. 12, 4313 (2021).
Budd, A. P. et al. Birth cohort effects in influenza surveillance data: evidence that first influenza infection affects later influenza-associated illness. J. Infect. Dis. 220, 820–829 (2019).
Fonville, J. M. et al. Antibody landscapes after influenza virus infection or vaccination. Science 346, 996–1000 (2014).
Lessler, J. et al. Evidence for antigenic seniority in influenza A (H3N2) antibody responses in southern China. PLoS Pathog. 8, e1002802 (2012).
Chen, R. & Holmes, E. C. The evolutionary dynamics of human influenza B virus. J. Mol. Evol. 66, 655–663 (2008).
Rosu, M. E. et al. Substitutions near the HA receptor binding site explain the origin and major antigenic change of the B/Victoria and B/Yamagata lineages. Proc. Natl Acad. Sci. USA 119, e2211616119 (2022).
Koutsakos, M., Nguyen, T. H., Barclay, W. S. & Kedzierska, K. Knowns and unknowns of influenza B viruses. Future Microbiol. 11, 119–135 (2016).
Skowronski, D. M. et al. Age-related differences in influenza B infection by lineage in a community-based sentinel system, 2010–2011 to 2015–2016, Canada. J. Infect. Dis. 216, 697–702 (2017).
Vijaykrishna, D. et al. The contrasting phylodynamics of human influenza B viruses. eLife 4, e05055 (2015).
Virk, R. K. et al. Divergent evolutionary trajectories of influenza B viruses underlie their contemporaneous epidemic activity. Proc. Natl Acad. Sci. USA 117, 619–628 (2020).
Yang, J. et al. Variation in influenza B virus epidemiology by lineage, China. Emerg. Infect. Dis. 24, 1536–1540 (2018).
Koutsakos, M., Wheatley, A. K., Laurie, K., Kent, S. J. & Rockman, S. Influenza lineage extinction during the COVID-19 pandemic? Nat. Rev. Microbiol. 19, 741–742 (2021).
Dhanasekaran, V. et al. Human seasonal influenza under COVID-19 and the potential consequences of influenza lineage elimination. Nat. Commun. 13, 1721 (2022).
Liu, Y. et al. Cross-lineage protection by human antibodies binding the influenza B hemagglutinin. Nat. Commun. 10, 324 (2019).
Dreyfus, C. et al. Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348 (2012).
Shen, C. et al. A multimechanistic antibody targeting the receptor binding site potently cross-protects against influenza B viruses. Sci. Transl. Med. 9, eaam5752 (2017).
Carlock, M. A. et al. Impact of age and pre-existing immunity on the induction of human antibody responses against influenza B viruses. Hum. Vaccin. Immunother. 15, 2030–2043 (2019).
Coudeville, L. et al. Relationship between haemagglutination-inhibiting antibody titres and clinical protection against influenza: development and application of a bayesian random-effects model. BMC Med. Res. Methodol. 10, 18 (2010).
Li, Y. et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J. Exp. Med. 210, 1493–1500 (2013).
Gouma, S. et al. Middle-aged individuals may be in a perpetual state of H3N2 influenza virus susceptibility. Nat. Commun. 11, 4566 (2020).
Linderman, S. L. et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013–2014 influenza season. Proc. Natl Acad. Sci. USA 111, 15798–15803 (2014).
Petrie, J. G. et al. Antibodies against the current influenza A(H1N1) vaccine strain do not protect some individuals from infection with contemporary circulating influenza A(H1N1) virus strains. J. Infect. Dis. 214, 1947–1951 (2016).
Flannery, B. et al. Influence of birth cohort on effectiveness of 2015–2016 influenza vaccine against medically attended illness due to 2009 pandemic influenza A(H1N1) virus in the United States. J. Infect. Dis. 218, 189–196 (2018).
Skowronski, D. M. et al. Beyond antigenic match: possible agent–host and immuno-epidemiological influences on influenza vaccine effectiveness during the 2015–2016 season in Canada. J. Infect. Dis. 216, 1487–1500 (2017).
Skowronski, D. M. et al. Paradoxical clade- and age-specific vaccine effectiveness during the 2018/19 influenza A(H3N2) epidemic in Canada: potential imprint-regulated effect of vaccine (I-REV). Euro Surveill. 24, 1900585 (2019).
Ranjeva, S. et al. Age-specific differences in the dynamics of protective immunity to influenza. Nat. Commun. 10, 1660 (2019).
Worobey, M., Plotkin, S. & Hensley, S. E. Influenza vaccines delivered in early childhood could turn antigenic sin into antigenic blessings. Cold Spring Harb. Perspect. Med. 10, a038471 (2020).
Bedford, T. et al. Integrating influenza antigenic dynamics with molecular evolution. eLife 3, e01914 (2014).
Langat, P. et al. Genome-wide evolutionary dynamics of influenza B viruses on a global scale. PLoS Pathog. 13, e1006749 (2017).
Wraith, S. et al. Homotypic protection against influenza in a pediatric cohort in Managua, Nicaragua. Nat. Commun. 13, 1190 (2022).
Skowronski, D. M. et al. Influenza B/Victoria antigen induces strong recall of B/Yamagata but lower B/Victoria response in children primed with two doses of B/Yamagata. Pediatr. Infect. Dis. J. 30, 833–839 (2011).
Lau, Y. C. et al. Variation by lineage in serum antibody responses to influenza B virus infections. PLoS ONE 15, e0241693 (2020).
Auladell, M. et al. Influenza virus infection history shapes antibody responses to influenza vaccination. Nat. Med. 28, 363–372 (2022).
Ertesvag, N. U. et al. Seasonal influenza vaccination expands hemagglutinin-specific antibody breadth to older and future A/H3N2 viruses. NPJ Vaccines 7, 67 (2022).
Kucharski, A. J., Lessler, J., Cummings, D. A. T. & Riley, S. Timescales of influenza A/H3N2 antibody dynamics. PLoS Biol. 16, e2004974 (2018).
Hensen, L., Kedzierska, K. & Koutsakos, M. Innate and adaptive immunity toward influenza B viruses. Future Microbiol. 15, 1045–1058 (2020).
Brady, R. C. et al. Randomized trial to compare the safety and immunogenicity of CSL Limited’s 2009 trivalent inactivated influenza vaccine to an established vaccine in United States children. Vaccine 32, 7141–7147 (2014).
Bodewes, R. et al. Prevalence of antibodies against seasonal influenza A and B viruses in children in Netherlands. Clin. Vaccine Immunol. 18, 469–476 (2011).
Sauerbrei, A. et al. Prevalence of antibodies against influenza A and B viruses in children in Germany, 2008 to 2010. Euro Surveill. 19, 20687 (2014).
Sauerbrei, A., Schmidt-Ott, R., Hoyer, H. & Wutzler, P. Seroprevalence of influenza A and B in German infants and adolescents. Med. Microbiol. Immunol. 198, 93–101 (2009).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012).
Rambaut, A., Lam, T. T., Max Carvalho, L. & Pybus, O. G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2, vew007 (2016).
Li, W. L. & Drummond, A. J. Model averaging and Bayes factor calculation of relaxed molecular clocks in Bayesian phylogenetics. Mol. Biol. Evol. 29, 751–761 (2012).
Shapiro, B., Rambaut, A. & Drummond, A. J. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 23, 7–9 (2006).
Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza (World Health Organization, 2011).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2016).
Edler, P. et al. Differential cross-reactivity to the influenza B virus haemagglutinin underpins lineage-specific susceptibility between birth cohorts. Zenodo https://doi.org/10.5281/zenodo.10633085 (2024).
Acknowledgements
We thank the participants for their generous involvement and provision of samples. We are grateful to the CSL investigators who originally initiated this study NCT00959049 trial. We are grateful to M. Vieira, K. Gostic and S. Cobey for discussions and advice on data analysis. This study has been generously supported by the Morningside Foundation and by Australian National Health and Medical Research Council Investigator grants (1195698 to M.K., 1173433 to A.K.W., 2009308 to J.A.J. and 1136322 to S.J.K.). The WHOCCRRI is supported by the Australian Government Department of Health. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper.
Author information
Authors and Affiliations
Contributions
M.K. designed and supervised the study. M.K., L.S.U.S., M.A. and N.S. performed the experiments. M.K., P.E., L.S.U.S., M.W., N.S., Y-M.D. and D.J.P. analysed data. M.A.C., T.M.R., J.A.J., S.R., S.J.K., A.K.W. and I.G.B. provided samples, reagents and/or data critical for the study. M.K., J.A.J., A.K.W., P.E. and D.J.P. contributed to the drafting of the paper. All authors reviewed the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
M.K. has acted as a consultant for Sanofi group of companies. S.R. is an employee of Seqirus, an influenza vaccine manufacturer. I.G.B. has shares in an influenza-vaccine-producing company. The other authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Sook-san Wong and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Cohort and dataset details.
(a) Details of the cohort associated with the main dataset generated for this study. The virus passage is indicated for each isolate (e – egg; c – cell). (b) Schematic of the relationships between year of virus isolation, birth year and sampling year. (c) Details of the cohort associated with the dataset generated by Carlock et al. (d) Details of the cohort associated with the WHOCCRI dataset.
Extended Data Fig. 2 Overview of HI titres.
(a) Correlations between antibody titres against the 19 IBV isolates from the main dataset (n = 322 serum samples). Virus isolates have been ordered by hierarchical clustering. (b) Antigenic characterization using human monoclonal antibodies against the IBV HA. End point titres from an HI assay are shown starting at 50 μg/ml. (c) Log2 HI titres against time (years) between sample collection and virus isolation (across all three datasets). (d) Log2 HI titres against time (years) between sample collection and virus isolation (across all three datasets) with individuals pooled into sample collection groups. For (C) and (D) the lines represent estimates using generalized additive models (GAMs) with 95% CI, accounting for repeated measurements on each individual by specifying a random effect. (e) Comparison of HI titres against cell and egg isolates in paired serums (n = 61) samples tested against cell or egg grown B/Phuket/2013 and B/Washington/2019. The number in blue indicated the difference in titres on the log2 scale. The boxplots show the median (centre line), quartiles (box limits) and maximum/minimum within 1.5x IQR above Q3 and below Q1(whiskers). P-values were generated from a Wilcoxon matched-pairs signed rank test (n = 61 samples from 2020). (f) Correlation between log2 HI titres against cell and egg grown B/Phuket/2013 and B/Washington/2019 viruses as described in C. (g) Correlation between log2 HI titres and microneutralization titres in 17 individuals tested against 10 viruses. In (F) and (G) the line represent estimates from linear regression with with 95% CI.
Extended Data Fig. 3 HI titres to past isolates against year of birth.
(a) Log2 HI titres to different isolates against year of birth separated by IBV lineage and dataset. The lines represent estimated mean HI titres from generalized additive models (GAMs). (b) Estimated mean HI titres against past isolates from each lineage that circulated prior to sample collection, against the year of birth for each participant. (c) Comparison of the three datasets for HI against each lineage by year of birth. The data is the same as in (B) but overlaid for three datasets per lineage. For (B) and (C) The lines represent the estimated mean HI titre from generalized additive models (GAMs) with shading representing 95% CIs, accounting for repeated measurements on each individual by specifying a random effect.
Extended Data Fig. 4 HI reactivity against past isolates by birth cohort in the main dataset.
Box-plots of antibody titres to specific IBV isolates for each birth cohort for the main dataset. P-values were generated from a Kruskal-Wallis test with Dunn’s correction for multiple comparisons. The boxplots show the median (centre line), quartiles (box limits) and maximum/minimum within 1.5x IQR above Q3 and below Q1(whiskers). The sample size for each birth cohort and virus is available in the Supplementary Table 3.
Extended Data Fig. 5 HI reactivity against past isolates by birth cohort in the supplementary datasets.
Box-plots of antibody titres to specific IBV isolates for each birth cohort for (a) the Carlock et al dataset and (b) the WHOCCRI dataset. P-values were generated from a Kruskal-Wallis test with Dunn’s correction for multiple comparisons. Only p-values > 0.05 are shown. The boxplots show the median (centre line), quartiles (box limits) and maximum/minimum within 1.5x IQR above Q3 and below Q1(whiskers). The sample size for each birth cohort and virus is available in the Supplementary Tables 4 and 5.
Extended Data Fig. 6 Antigenic seniority in IBV HI titres.
(a) Distribution of the age of the participant at the time of isolation of the strain to which they had highest antibody titres, grouped by IBV lineage or birth cohort. (b) HI titres relative to the age of the participant at the time of virus isolation. (c) HI titres relative to the age of the participant at the time of sampling. For (B) and (C) the lines represent estimated mean HI titres from generalized additive models (GAMs) with shaded region representing 95% CIs, accounting for repeated measurements on each individual by specifying a random effect. Only titrations of viruses isolated prior to sample collection are included. Analysis shown for measurements from the main (A, B, C) or Carclock et al. (B).
Extended Data Fig. 7 HI titres to future isolates by birth year in supplementary datasets.
(a) Estimated mean HI titres against the future unencountered B/Colorado/02/2017 from the Carlock et al dataset. The lines represent estimated mean HI titres from generalized additive models (GAMs) with shaded region representing 95% CIs. (b) Box-plots of HI titres to future unencountered B/Colorado/02/2017 for each birth cohort from the Carlock et al dataset. The boxplots show the median (centre line), quartiles (box limits) and maximum/minimum within 1.5x IQR above Q3 and below Q1(whiskers). with n = 5 (1917–1939), n = 35 (1940–1960), n = 26 (1961–1980), n = 19 (1981–1998).
Extended Data Fig. 8 Estimated susceptibility to future isolates varies by birth year.
Sensitivity analysis of estimated HI (a, b) and probabilities of infection (c, d) by birth year for the effects of different virus isolates (A,C) or sampling year groups (B,D). The median estimated HI or probability is shown and the shaded areas represent the 25th and 75th percentiles. Data from the main dataset were used.
Supplementary information
Supplementary Information
Supplementary information guide.
Supplementary Tables
Supplementary Tables 1–7.
Source data
Source Data Fig. 1
Sequence data and statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Edler, P., Schwab, L.S.U., Aban, M. et al. Immune imprinting in early life shapes cross-reactivity to influenza B virus haemagglutinin. Nat Microbiol 9, 2073–2083 (2024). https://doi.org/10.1038/s41564-024-01732-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41564-024-01732-8
This article is cited by
-
Influenza vaccination post-COVID-19 expands vaccine-specific effector CD4 T-cells and Tregs under positive influence of host trained innate immunity
npj Vaccines (2025)
-
Immunological sin: how a person’s earliest flu infections dictate life-long immunity
Nature (2025)
-
Immunological imprinting and risks of influenza B virus infection
Nature Immunology (2024)
-
Preclinical evaluation of a universal inactivated influenza B vaccine based on the mosaic hemagglutinin-approach
npj Vaccines (2024)