Abstract
Ribosomes translate mRNA into protein. Despite divergence in ribosome structure over the course of evolution, the catalytic site, known as the peptidyl transferase centre (PTC), is thought to be nearly universally conserved. Here we identify clades of archaea that have highly divergent ribosomal RNA sequences in the PTC. To understand how these PTC sequences fold, we determined cryo-EM structures of the 70S and 50S ribosomes to 2.4 Å and 2 Å, respectively, from the hyperthermophilic archaeon Pyrobaculum calidifontis. PTC sequence variation leads to the rearrangement of key base triples, and differences between archaeal and bacterial ribosomal proteins enable sequence variation in archaeal PTCs. Finally, we identify an archaeal ribosome hibernation factor, Dri, that differs from known bacterial and eukaryotic hibernation factors and is found in multiple archaeal phyla. Overall, this work identifies factors that regulate ribosome function in archaea and reveals a larger diversity of the most ancient sequences in the ribosome.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Supporting data are available within the Article and its Supplementary Information. Ribosome coordinates have been deposited in the Protein Data Bank for the P. calidifontis 50S-Dri complex (9E6Q), 70S consensus reconstruction (9E71) and 70S-Dri complex (9E7F). Cryo-EM maps have been deposited in the Electron Microscopy Data Bank for the 50S-Dri complex (EMD-47578), 70S consensus reconstruction (EMD-47604, EMD-47605, EMD-47606, EMD-47611, EMD-47617 and EMD-47628) and 70S-Dri complex (EMD-47662, EMD-47664, EMD-47666, EMD-47667 and EMD-47668). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD064958. Ribosome structures from the Protein Data Bank (PBD IDs 1S72, 1VY4, 6SKF, 7K00 and 8EMM) were used in this study. Source data are provided with this paper.
References
Petrov, A. S. et al. History of the ribosome and the origin of translation. Proc. Natl Acad. Sci. USA 112, 15396–15401 (2015).
Tirumalai, M. R., Rivas, M., Tran, Q. & Fox, G. E. The peptidyl transferase center: a window to the past. Microbiol. Mol. Biol. Rev. 85, e0010421 (2021).
Noller, H. F., Donohue, J. P. & Gutell, R. R. The universally conserved nucleotides of the small subunit ribosomal RNAs. RNA 28, 623–644 (2022).
Noeske, J. et al. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 22, 336–341 (2015).
Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 (2000).
Voorhees, R. M., Schmeing, T. M., Kelley, A. C. & Ramakrishnan, V. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330, 835–838 (2010).
Harms, J. et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107, 679–688 (2001).
Ben-Shem, A., Jenner, L., Yusupova, G. & Yusupov, M. Crystal structure of the eukaryotic ribosome. Science 330, 1203–1209 (2010).
Fromm, S. A. et al. The translating bacterial ribosome at 1.55 Å resolution generated by cryo-EM imaging services. Nat. Commun. 14, 1095 (2023).
Smirnova, J. et al. Structure of the actively translating plant 80S ribosome at 2.2 Å resolution. Nat. Plants 9, 987–1000 (2023).
Holm, M. et al. mRNA decoding in human is kinetically and structurally distinct from bacteria. Nature 617, 200–207 (2023).
Eiler, D. R. et al. The Giardia lamblia ribosome structure reveals divergence in several biological pathways and the mode of emetine function. Structure 32, 400–410 (2024).
Helena-Bueno, K. et al. A new family of bacterial ribosome hibernation factors. Nature 626, 1125–1132 (2024).
Perez Boerema, A. et al. Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nat. Plants 4, 212–217 (2018).
Singh, V. et al. Mitoribosome structure with cofactors and modifications reveals mechanism of ligand binding and interactions with L1 stalk. Nat. Commun. 15, 4272 (2024).
Harper, N. J., Burnside, C. & Klinge, S. Principles of mitoribosomal small subunit assembly in eukaryotes. Nature 614, 175–181 (2023).
Borrel, G., Brugère, J.-F., Gribaldo, S., Schmitz, R. A. & Moissl-Eichinger, C. The host-associated archaeome. Nat. Rev. Microbiol. 18, 622–636 (2020).
Cai, M. et al. Ecological features and global distribution of Asgard archaea. Sci. Total Environ. 758, 143581 (2021).
Valentin-Alvarado, L. E. et al. Asgard archaea modulate potential methanogenesis substrates in wetland soil. Nat. Commun. 15, 6384 (2024).
Brochier-Armanet, C., Boussau, B., Gribaldo, S. & Forterre, P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6, 245–252 (2008).
Kohtz, A. J. et al. Cultivation and visualization of a methanogen of the phylum Thermoproteota. Nature 632, 1118–1123 (2024).
Sas-Chen, A. et al. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature 583, 638–643 (2020).
Ohira, T. et al. Reversible RNA phosphorylation stabilizes tRNA for cellular thermotolerance. Nature 605, 372–379 (2022).
van den Elzen, A., Helena-Bueno, K., Brown, C. R., Chan, L. I. & Melnikov, S. V. Ribosomal proteins can hold a more accurate record of bacterial thermal adaptation compared to rRNA. Nucleic Acids Res. 51, 8048–8059 (2023).
Nissley, A. J., Penev, P. I., Watson, Z. L., Banfield, J. F. & Cate, J. H. D. Rare ribosomal RNA sequences from archaea stabilize the bacterial ribosome. Nucleic Acids Res. 51, 1880–1894 (2023).
Nürenberg-Goloub, E. et al. Molecular analysis of the ribosome recycling factor ABCE1 bound to the 30S post-splitting complex. EMBO J. 39, e103788 (2020).
Kazan, R. et al. Role of aIF5B in archaeal translation initiation. Nucleic Acids Res. 50, 6532–6548 (2022).
Wang, Y.-H. et al. Cryo-electron microscopy structure and translocation mechanism of the crenarchaeal ribosome. Nucleic Acids Res. 51, 8909–8924 (2023).
Doris, S. M. et al. Universal and domain-specific sequences in 23S–28S ribosomal RNA identified by computational phylogenetics. RNA 21, 1719–1730 (2015).
Polacek, N. & Mankin, A. S. The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit. Rev. Biochem. Mol. Biol. 40, 285–311 (2005).
Ishida, S., Ngo, P. H. T., Gundlach, A. & Ellington, A. Engineering ribosomal machinery for noncanonical amino acid incorporation. Chem. Rev. 124, 7712–7730 (2024).
d’Aquino, A. E., Kim, D. S. & Jewett, M. C. Engineered ribosomes for basic science and synthetic biology. Annu. Rev. Chem. Biomol. Eng. 9, 311–340 (2018).
Kjems, J. et al. Gene organization, transcription signals and processing of the single ribosomal RNA operon of the archaebacterium Thermoproteus tenax. Nucleic Acids Res. 15, 4821–4835 (1987).
Amo, T. et al. Pyrobaculum calidifontis sp. nov., a novel hyperthermophilic archaeon that grows in atmospheric air. Archaea 1, 113–121 (2002).
Reeve, C. A., Amy, P. S. & Matin, A. Role of protein synthesis in the survival of carbon-starved Escherichia coli K-12. J. Bacteriol. 160, 1041–1046 (1984).
Kolter, R., Siegele, D. A. & Tormo, A. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47, 855–874 (1993).
Prossliner, T., Gerdes, K., Sørensen, M. A. & Winther, K. S. Hibernation factors directly block ribonucleases from entering the ribosome in response to starvation. Nucleic Acids Res. 49, 2226–2239 (2021).
Helena-Bueno, K., Chan, L. I. & Melnikov, S. V. Rippling life on a dormant planet: hibernation of ribosomes, RNA polymerases, and other essential enzymes. Front. Microbiol. 15, 1386179 (2024).
Prossliner, T., Skovbo Winther, K., Sørensen, M. A. & Gerdes, K. Ribosome hibernation. Annu. Rev. Genet. 52, 321–348 (2018).
Londei, P., Altamura, S., Caprini, E. & Martayan, A. Translation and ribosome assembly in extremely thermophilic archaebacteria. Biochimie 73, 1465–1472 (1991).
Armache, J.-P. et al. Promiscuous behaviour of archaeal ribosomal proteins: implications for eukaryotic ribosome evolution. Nucleic Acids Res. 41, 1284–1293 (2013).
Coureux, P.-D., Lazennec-Schurdevin, C., Bourcier, S., Mechulam, Y. & Schmitt, E. Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation. Commun. Biol. 3, 58 (2020).
Jamali, K. et al. Automated model building and protein identification in cryo-EM maps. Nature 628, 450–457 (2024).
Baker, B. J. et al. Enigmatic, ultrasmall, uncultivated Archaea. Proc. Natl Acad. Sci. USA 107, 8806–8811 (2010).
Klein, D. J., Moore, P. B. & Steitz, T. A. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 340, 141–177 (2004).
Thompson, J. et al. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc. Natl Acad. Sci. USA 98, 9002–9007 (2001).
d’Aquino, A. E. et al. Mutational characterization and mapping of the 70S ribosome active site. Nucleic Acids Res. 48, 2777–2789 (2020).
Sato, N. S., Hirabayashi, N., Agmon, I., Yonath, A. & Suzuki, T. Comprehensive genetic selection revealed essential bases in the peptidyl-transferase center. Proc. Natl Acad. Sci. USA 103, 15386–15391 (2006).
Blaha, G., Gürel, G., Schroeder, S. J., Moore, P. B. & Steitz, T. A. Mutations outside the anisomycin-binding site can make ribosomes drug-resistant. J. Mol. Biol. 379, 505–519 (2008).
Kloss, P., Xiong, L., Shinabarger, D. L. & Mankin, A. S. Resistance mutations in 23 S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J. Mol. Biol. 294, 93–101 (1999).
Watson, Z. L. et al. Structure of the bacterial ribosome at 2 Å resolution. eLife 9, e60482 (2020).
Lhoest, J. & Colson, C. Genetics of ribosomal protein methylation in Escherichia coli. II. A mutant lacking a new type of methylated amino acid, N5-methylglutamine, in protein L3. Mol. Gen. Genet. 154, 175–180 (1977).
Li, Q. I. et al. A modified pCas/pTargetF system for CRISPR–Cas9-assisted genome editing in Escherichia coli. Acta Biochim. Biophys. Sin. 53, 620–627 (2021).
Bateman, A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 22, 12–13 (1997).
Watson, Z. L. et al. Atomistic simulations of the Escherichia coli ribosome provide selection criteria for translationally active substrates. Nat. Chem. 15, 913–921 (2023).
Polikanov, Y. S., Steitz, T. A. & Innis, C. A. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 21, 787–793 (2014).
Schmeing, T. M., Huang, K. S., Strobel, S. A. & Steitz, T. A. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438, 520–524 (2005).
Budkevich, T. et al. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol. Cell 44, 214–224 (2011).
Budkevich, T. V. et al. Regulation of the mammalian elongation cycle by subunit rolling: a eukaryotic-specific ribosome rearrangement. Cell 158, 121–131 (2014).
Yusupov, M. M. et al. Crystal structure of the ribosome at 5.5 A resolution. Science 292, 883–896 (2001).
Dixon, A. S. et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 11, 400–408 (2016).
Londei, P., Altamura, S., Cammarano, P. & Petrucci, L. Differential features of ribosomes and of poly(U)‐programmed cell‐free systems derived from sulphur‐dependent archaebacterial species. Eur. J. Biochem. 157, 455–462 (1986).
Vila-Sanjurjo, A., Schuwirth, B.-S., Hau, C. W. & Cate, J. H. D. Structural basis for the control of translation initiation during stress. Nat. Struct. Mol. Biol. 11, 1054–1059 (2004).
Hummel, H. & Böck, A. 23S ribosomal RNA mutations in halobacteria conferring resistance to the anti-80S ribosome targeted antibiotic anisomycin. Nucleic Acids Res. 15, 2431–2443 (1987).
Kearsey, S. E. & Craig, I. W. Altered ribosomal RNA genes in mitochondria from mammalian cells with chloramphenicol resistance. Nature 290, 607–608 (1981).
Mailliot, J. et al. Crystal structures of the uL3 mutant ribosome: illustration of the importance of ribosomal proteins for translation efficiency. J. Mol. Biol. 428, 2195–2202 (2016).
Ward, F. R., Watson, Z. L., Ad, O., Schepartz, A. & Cate, J. H. D. Defects in the assembly of ribosomes selected for β-amino acid incorporation. Biochemistry 58, 4494–4504 (2019).
Radford, F., Rinehart, J. & Isaacs, F. J. Mapping the in vivo fitness landscape of a tethered ribosome. Sci. Adv. 9, eade8934 (2023).
Yaeshima, C. et al. A novel ribosome-dimerization protein found in the hyperthermophilic archaeon Pyrococcus furiosus using ribosome-associated proteomics. Biochem. Biophys. Res. Commun. 593, 116–121 (2022).
Hassan, A. H. et al. Novel archaeal ribosome dimerization factor facilitating unique 30S–30S dimerization. Nucleic Acids Res. 53, gkae1324 (2025).
Polikanov, Y. S., Blaha, G. M. & Steitz, T. A. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336, 915–918 (2012).
Barandun, J., Hunziker, M., Vossbrinck, C. R. & Klinge, S. Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome. Nat. Microbiol. 4, 1798–1804 (2019).
Schorb, M., Haberbosch, I., Hagen, W. J. H., Schwab, Y. & Mastronarde, D. N. Software tools for automated transmission electron microscopy. Nat. Methods 16, 471–477 (2019).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Kimanius, D., Dong, L., Sharov, G., Nakane, T. & Scheres, S. H. W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 478, 4169–4185 (2021).
Casañal, A., Lohkamp, B. & Emsley, P. Current developments in Coot for macromolecular model building of electron cryo‐microscopy and crystallographic data. Protein Sci. 29, 1055–1064 (2020).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019).
Parks, D. H. et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 50, D785–D794 (2022).
Nawrocki, E. P. & Eddy, S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013).
Kalvari, I. et al. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 49, D192–D200 (2021).
Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Penev, P. I. et al. ProteoVision: web server for advanced visualization of ribosomal proteins. Nucleic Acids Res. 49, W578–W588 (2021).
Eddy, S. R. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 (2011).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
UniProt Consortium. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531 (2023).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82 (2024).
Sowers, K. R., Boone, J. E. & Gunsalus, R. P. Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Appl. Environ. Microbiol. 59, 3832–3839 (1993).
Liu, H., Lin, D. & Yates, J. R. Multidimensional separations for protein/peptide analysis in the post-genomic era. BioTechniques 32, 898–911 (2002).
Washburn, M. P., Wolters, D. & Yates, J. R. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 (2001).
Perez-Riverol, Y. et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 53, D543–D553 (2025).
Douthwaite, S., Powers, T., Lee, J. Y. & Noller, H. F. Defining the structural requirements for a helix in 23S ribosomal RNA that confers erythromycin resistance. J. Mol. Biol. 209, 655–665 (1989).
Nissley, A. J., Kamal, T. S. & Cate, J. H. D. Interactions between terminal ribosomal RNA helices stabilize the E. coli large ribosomal subunit. RNA 29, 1500–1508 (2023).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Acknowledgements
We thank D. Toso, R. Thakkar and P. Tobias for assistance with cryo-EM data collection (Cal-Cryo). This work was funded by the NSF Center for Genetically Encoded Materials (C-GEM) (CHE-2002182). Y.S. is a Don Brown Awardee of the Life Sciences Research Foundation. D.D.N. is a Chan Zuckerberg Biohub—San Francisco Investigator. This work used the Vincent J. Coates Proteomics/Mass Spectrometry Laboratory Core Facility, RRID: SCR_025852.
Author information
Authors and Affiliations
Contributions
A.J.N., P.I.P. and J.H.D.C. conceptualized the project. A.J.N. and B.E.D. cultured archaea. A.J.N. acquired cryo-EM data and conducted image analysis and modelling. A.J.N. and R.W.K. performed biochemical experiments. Y.S. performed bioinformatic analysis. A.J.N. and Y.S. prepared figures. A.J.N. wrote the initial draft, and all authors reviewed and edited the paper. J.H.D.C., J.F.B. and D.D.N. were responsible for funding and supervised the project.
Corresponding author
Ethics declarations
Competing interests
J.H.D.C. is a founder and board and SAB member of Initial Therapeutics. The other authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Sergey Melnikov, Thomas Santangelo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
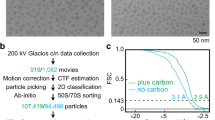
Extended Data Fig. 1 Cryo-EM sample preparation and analysis of post-transcriptional modifications in P. calidifontis.
a, Sucrose gradient fractionation of P. calidifontis ribosomes. Dashed lines bracket the sample that was collected for cryo-EM analysis. b, Distribution of archaea-specific rProteins across four orders and further subclades in the class Thermoprotei. Closed and open circles represent clades where rProtein homologs were or were not identified, respectively. c, Zinc binding sites in LSU rProtein eL37 and SSU rProtein uS17 with cryo-EM density. d-e, Cryo-EM density for representative post-transcriptional and post-translational modifications in the LSU (d) and SSU (e). f, Distribution of post-transcriptional modifications in P. calidifontis (left) and T. kodakarensis PDB:6SKF ref. 22 (right). Note that pseudouridines are not modeled in T. kodakarensis.
Extended Data Fig. 2 Sequence and structural diversity in archaeal PTCs.
a, Shannon entropy plot for PTC nucleotides in GTDB bacteria and archaea. Positions indicated by a * have a Shannon entropy value of zero for archaea. A Shannon entropy value of zero corresponds to complete conservation of the nucleotide, while a value of 1.386 corresponds to an equal distribution of all four nucleotides. b, Distribution of rare PTC sequences in Nanoarchaeota archaea. The cladogram is derived from the GTDB phylogeny, where branches with bootstrap support <50% are colored in grey. The letter o represents order and n represents the number of 23S rRNA sequences in each distribution. Nucleotides that differ from the archaeal consensus sequence are shown in color. c-e, Cryo-EM density for regions of the PTC shown in Fig. 2. Water molecules and magnesium ions are shown as red and green spheres respectively. f, Secondary structures of the H. marismortui and P. calidifontis A loops. Positions with rare sequence variation, 2554 and 2555, are highlighted in red in the P. calidifontis A loop. g, Comparison of the apical region of the A loop in H. marismortui PDB:1S72 ref. 45 (blue) and P. calidifontis (purple). h, Cryo-EM density for the P. calidifontis A loop. Base stacking interactions are shown as dashed lines.
Extended Data Fig. 3 Differences in uL3 and the PTC between the domains of life.
a, Amino acid occupancy in an alignment of uL3 sequences from representative species in bacteria, archaea, and eukaryotes84. b, Sequencing chromatogram for the rplC gene from ∆Q150-uL3 E. coli, showing the deletion of residue Q150. The chromatogram trace was broken at the site of deletion for clarity. c, Comparison of the positions of 23S rRNA nucleotides 2452 and 2504 in E. coli and archaeal ribosomes. d, RT-PCR purity assay for MS2-tagged E. coli ribosomes used in this study. Sample purity was quantified using the band intensities of the upper (MS2-tagged ribosome) and lower (WT ribosome) bands.
Extended Data Fig. 4 Dri interactions with the ribosome.
a, Dri (red) binds to the rotated state of the P. calidifontis ribosome (grey). An E. coli ribosome in the unrotated state (PDB:8EMM ref. 55) is overlayed for reference (grey). b, (left) Dri residues occupy the positions of the A and P-site amino acids in the PTC. The A and P-site tRNAs from PDB:8EMM ref. 55 are overlayed on the P. calidifontis ribosome (right). Dri residue F219 occupies the A-site cleft in a position similar to phenylalanyl-tRNAPhe (PDB: 1VY4 ref. 56). c, In the absence of the Dri protein, a loop of rProtein uL16 is disordered (top). Cryo-EM density is shown in gray. Upon binding of the Dri N-terminal lobe to the LSU, the loop of uL16 becomes ordered (bottom). d, Cryo-EM density for PTC nucleotides in the 50S-Dri complex (left) or the composite 70S complex (right).
Extended Data Fig. 5 Dri expression and association with the ribosome.
a, Recombinantly expressed and purified Dri proteins resolved on a 4–12% polyacrylamide gel stained with Coomassie brilliant blue. b, P. calidifontis lysate-based in vitro translation assay. After translation of the HiBiT peptide, luminescence was measured from the HiBiT-LgBiT complex. For the no mRNA and buffer controls, 3 µL of Dri buffer A was added to the reaction. Each point on the graph represents an individual measurement, and data and error bars represent the mean and standard deviation of the three measurements, respectively. The fold change from the buffer control is indicated on top of each bar. A two-sided, two-sample t-test was used to compare values (p-values from left to right: 6x10−5, 3x10−6, 7x10−8, 4x10−8, 2x10−7, 4x10−8, 2x10−4, 9x10−5, 7x10−8). c, Sucrose gradient fractionation of ribosomes from P. calidifontis cultures grown to the indicated optical densities. Data was overlayed based on the 50S peak and grey lines bracket the sample that was collected for mass spectrometry. d, Fold change, measured by mass spectrometry, in protein levels from P. calidifontis 70S ribosomes samples isolated from stationary and logarithmic phase cultures. P-values were calculated with ANOVA in PEAKS Studio. e, The difference in Dri and uL2 mRNA Ct values from total RNA extracted from cultures at logarithmic (OD600 0.07) or stationary (OD600 0.15) phase, measured by RT-qPCR. Each point on the graph represents an individual measurement, and data and error bars represent the mean and standard deviation of the three measurements, respectively. A two-sided, two-sample t-test was used to compare values (p-value=0.046). f, Sucrose gradient fractionation of M. acetivorans ribosomes. Grey lines bracket the sample that was collected for mass spectrometry.
Supplementary information
Supplementary Information
Supplementary Notes 1–3, Figs. 1–7 and Tables 1–10.
Supplementary Data 1
23S rRNA sequences from GTDB archaea.
Supplementary Data 2
23S rRNA sequences from GTDB bacteria.
Supplementary Data 3
PTC sequence conservation, distribution of archaeal rProteins and Dri homologues, and all proteins identified by mass spectrometry in this study.
Supplementary Data 4
Sequences of rProtein aL48 homologues.
Supplementary Data 5
Sequences of rProtein aL49 homologues.
Supplementary Data 6
Sequences of rProtein aS21 homologues.
Supplementary Data 7
Sequences of rProtein aS35 homologues.
Supplementary Data 8
Sequences of Dri homologues that contain both the N- and C-terminal lobes.
Supplementary Data 9
Sequences of Dri homologues that contain only the N-terminal lobe.
Supplementary Data 10
Sequences of Dri homologues that contain only the C-terminal lobe.
Source data
Source Data Figs. 3 and 5 and Extended Data Figs. 1–3 and 5
Statistical source data.
Source Data Extended Data Figs. 3 and 5
Uncropped gels.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nissley, A.J., Shulgina, Y., Kivimae, R.W. et al. Structure of an archaeal ribosome reveals a divergent active site and hibernation factor. Nat Microbiol 10, 1940–1953 (2025). https://doi.org/10.1038/s41564-025-02065-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41564-025-02065-w
This article is cited by
-
Circularization of 23S rRNA but not 16S rRNA within archaeal ribosomes
Genome Biology (2026)