Abstract
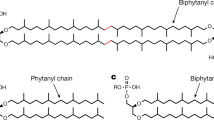
Bathyarchaeia, an abundant and ecologically versatile archaea found commonly in marine sediments, has a key role in the global carbon cycle. However, its lipid biomarkers and carbon assimilation mechanisms are poorly understood. Here, using a highly enriched Bathyarchaeia culture (>95% archaea) obtained from estuarine sediment of the East China Sea, we show that Baizosediminiarchaeum (formerly subgroup Bathy-8), the most abundant and widespread Bathyarchaeia group on Earth, synthesizes butanetriol dialkyl glycerol tetraethers (BDGTs) as its dominant membrane lipids. BDGTs are unusual archaeal tetraether lipids characterized by a butanetriol backbone instead of the typical glycerol, challenging fundamental assumptions in archaeal lipid biochemistry. Although BDGTs have been previously identified in the methanogen Methanomassiliicoccus luminyensis, we now provide direct evidence that Bathyarchaeia also synthesizes BDGTs, definitively establishing this globally abundant group as a natural BDGT producer. Stable isotope probing with 13C-bicarbonate shows that Baizosediminiarchaeum assimilates carbon into BDGTs from both inorganic carbon and lignin. These unique carbon assimilation strategies suggest the biogeochemical importance of Baizosediminarchaeum in marine carbon cycling and organic matter decomposition.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The 16S rRNA gene sequencing data from the East China Sea (Fig. 1) have been deposited in the NCBI SRA under accession number PRJNA1303662 and in the National Omics Data Encyclopedia under accession number OEP00006462. Mass spectrometry data supporting Fig. 2 and Extended Data Fig. 2 are publicly available via Zenodo at https://doi.org/10.5281/zenodo.16742269 (ref. 63). Stable isotope data supporting Fig. 4 are available via Zenodo at https://doi.org/10.5281/zenodo.16748030 (ref. 64). Source data are provided with this paper.
References
Meng, J. et al. Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. ISME J. 8, 650–659 (2014).
He, Y. et al. Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat. Microbiol. 1, 16035 (2016).
Hou, J. et al. Taxonomic and carbon metabolic diversification of Bathyarchaeia during its coevolution history with early Earth surface environment. Sci. Adv. 9, eadf5069 (2023).
Rinke, C. et al. A standardized archaeal taxonomy for the Genome Taxonomy Database. Nat. Microbiol. 6, 946–959 (2021).
Feng, X., Wang, Y., Zubin, R. & Wang, F. Core metabolic features and hot origin of Bathyarchaeota. Engineering 5, 498–504 (2019).
Lloyd, K. G. et al. Predominant archaea in marine sediments degrade detrital proteins. Nature 496, 215–218 (2013).
Yu, T. et al. Growth of sedimentary Bathyarchaeota on lignin as an energy source. Proc. Natl Acad. Sci. USA 115, 6022–6027 (2018).
Lazar, C. S. et al. Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environ. Microbiol. 18, 1200–1211 (2016).
Evans, P. N. et al. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350, 434–438 (2015).
Wang, Y., Wegener, G., Ruff, S. E. & Wang, F. Methyl/alkyl‐coenzyme M reductase‐based anaerobic alkane oxidation in archaea. Environ. Microbiol. 23, 530–541 (2021).
Yu, T. et al. Widespread Bathyarchaeia encode a novel methyltransferase utilizing lignin‐derived aromatics. mLife 2, 272–282 (2023).
Khomyakova, M. A., Merkel, A. Y., Mamiy, D. D., Klyukina, A. A. & Slobodkin, A. I. Phenotypic and genomic characterization of Bathyarchaeum tardum gen. nov., sp. nov., a cultivated representative of the archaeal class Bathyarchaeia. Front. Microbiol. 14, 1214631 (2023).
Buckles, L. K., Villanueva, L., Weijers, J. W., Verschuren, D. & Damsté, J. S. S. Linking isoprenoidal GDGT membrane lipid distributions with gene abundances of ammonia‐oxidizing Thaumarchaeota and uncultured crenarchaeotal groups in the water column of a tropical lake (Lake Challa, East Africa). Environ. Microbiol. 15, 2445–2462 (2013).
Coffinet, S. et al. Structural elucidation and environmental distributions of butanetriol and pentanetriol dialkyl glycerol tetraethers (BDGTs and PDGTs). Biogeosciences 17, 317–330 (2020).
Meador, T. B. et al. The archaeal lipidome in estuarine sediment dominated by members of the Miscellaneous Crenarchaeotal Group. Environ. Microbiol. 17, 2441–2458 (2015).
Zhu, C., Meador, T. B., Dummann, W. & Hinrichs, K. U. Identification of unusual butanetriol dialkyl glycerol tetraether and pentanetriol dialkyl glycerol tetraether lipids in marine sediments. Rapid Commun. Mass Spectrom. 28, 332–338 (2014).
Becker, K. W. et al. Unusual butane- and pentanetriol-based tetraether lipids in Methanomassiliicoccus luminyensis, a representative of the seventh order of methanogens. Appl. Environ. Microbiol. 82, 4505–4516 (2016).
Janssen, P. H. & Kirs, M. Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 74, 3619–3625 (2008).
Paul, K., Nonoh, J. O., Mikulski, L. & Brune, A. “Methanoplasmatales,” Thermoplasmatales-related archaea in termite guts and other environments, are the seventh order of methanogens. Appl. Environ. Microbiol. 78, 8245–8253 (2012).
Poulsen, M. et al. Methylotrophic methanogenic Thermoplasmata implicated in reduced methane emissions from bovine rumen. Nat. Commun. 4, 1428 (2013).
Fillol, M., Auguet, J.-C., Casamayor, E. O. & Borrego, C. M. Insights in the ecology and evolutionary history of the Miscellaneous Crenarchaeotic Group lineage. ISME J. 10, 665–677 (2016).
Söllinger, A. et al. Phylogenetic and genomic analysis of Methanomassiliicoccales in wetlands and animal intestinal tracts reveals clade-specific habitat preferences. FEMS Microbiol. Ecol. 92, fiv149 (2016).
Zhou, Z., Chen, J., Cao, H., Han, P. & Gu, J.-D. Analysis of methane-producing and metabolizing archaeal and bacterial communities in sediments of the northern South China Sea and coastal Mai Po Nature Reserve revealed by PCR amplification of mcrA and pmoA genes. Front. Microbiol. 5, 789 (2015).
Lazar, C. S. et al. Environmental controls on intragroup diversity of the uncultured benthic archaea of the miscellaneous Crenarchaeotal group lineage naturally enriched in anoxic sediments of the White Oak River estuary (North Carolina, USA). Environ. Microbiol. 17, 2228–2238 (2015).
Kubo, K. et al. Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. ISME J. 6, 1949–1965 (2012).
Hoshino, T. et al. Global diversity of microbial communities in marine sediment. Proc. Natl Acad. Sci. USA 117, 27587–27597 (2020).
Rubin-Blum, M. et al. Diversity, activity, and abundance of benthic microbes in the Southeastern Mediterranean Sea. FEMS Microbiol. Ecol. 98, fiac009 (2022).
Zhang, C. et al. Marine sediments harbor diverse archaea and bacteria with the potential for anaerobic hydrocarbon degradation via fumarate addition. FEMS Microbiol. Ecol. 97, fiab045 (2021).
Elling, F. J. et al. Effects of growth phase on the membrane lipid composition of the thaumarchaeon Nitrosopumilus maritimus and their implications for archaeal lipid distributions in the marine environment. Geochim. Cosmochim. Acta 141, 579–597 (2014).
Simon, M. & Azam, F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser. 51, 201–213 (1989).
Blewett, J., Naafs, B., Gallego-Sala, A. & Pancost, R. D. Effects of temperature and pH on archaeal membrane lipid distributions in freshwater wetlands. Org. Geochem. 148, 104080 (2020).
Takano, Y. et al. Sedimentary membrane lipids recycled by deep-sea benthic archaea. Nat. Geosci. 3, 858–861 (2010).
Knappy, C. et al. Mono‐, di‐ and trimethylated homologues of isoprenoid tetraether lipid cores in archaea and environmental samples: mass spectrometric identification and significance. J. Mass Spectrom. 50, 1420–1432 (2015).
Elling, F. J. et al. Chemotaxonomic characterisation of the thaumarchaeal lipidome. Environ. Microbiol. 19, 2681–2700 (2017).
Meador, T. B. et al. Thermococcus kodakarensis modulates its polar membrane lipids and elemental composition according to growth stage and phosphate availability. Front. Microbiol. 5, 10 (2014).
Niederberger, T. D., Götz, D. K., McDonald, I. R., Ronimus, R. S. & Morgan, H. W. Ignisphaera aggregans gen. nov., sp. nov., a novel hyperthermophilic crenarchaeote isolated from hot springs in Rotorua and Tokaanu, New Zealand. Int. J. Syst. Evol. Microbiol. 56, 965–971 (2006).
Zhu, Q.-Z., Elvert, M., Meador, T. B., Becker, K. W. & Heuer, V. B. Stable carbon isotopic compositions of archaeal lipids constrain terrestrial, planktonic, and benthic sources in marine sediments. Geochim. Cosmochim. Acta 307, 319–337 (2021).
Gliozzi, A., Relini, A. & Chong, P. L.-G. Structure and permeability properties of biomimetic membranes of bolaform archaeal tetraether lipids. J. Membr. Sci. 206, 131–147 (2002).
Blewett, J. et al. Metabolic and ecological controls on the stable carbon isotopic composition of archaeal (isoGDGT and BDGT) and bacterial (brGDGT) lipids in wetlands and lignites. Geochim. Cosmochim. Acta 320, 1–25 (2022).
Liang, W., Yu, T., Dong, L., Jia, Z. & Wang, F. Determination of carbon-fixing potential of Bathyarchaeota in marine sediment by DNA stable isotope probing analysis. Sci. China Earth Sci. 66, 910–917 (2023).
Yin, X. et al. Physiological versatility of ANME-1 and Bathyarchaeotoa-8 archaea evidenced by inverse stable isotope labeling. Microbiome 12, 68 (2024).
Jain, S., Caforio, A. & Driessen, A. J. Biosynthesis of archaeal membrane ether lipids. Front. Microbiol. 5, 641 (2014).
Londry, K. L., Dawson, K. G., Grover, H. D., Summons, R. E. & Bradley, A. S. Stable carbon isotope fractionation between substrates and products of Methanosarcina barkeri. Org. Geochem. 39, 608–621 (2008).
Wu, W. et al. Substrate‐dependent incorporation of carbon and hydrogen for lipid biosynthesis by Methanosarcina barkeri. Environ. Microbiol. Rep. 12, 555–567 (2020).
Hayes, J. M. Fractionation of carbon and hydrogen isotopes in biosynthetic processes. in Stable Isotope Geochemistry 1st edn (eds Valley, J. W. & Cole, D. R.) Ch. 3 (De Gruyter, 2018).
Kurth, J. M. et al. Methanogenic archaea use a bacteria-like methyltransferase system to demethoxylate aromatic compounds. ISME J. 15, 3549–3565 (2021).
Lloyd, M. et al. Methoxyl stable isotopic constraints on the origins and limits of coal-bed methane. Science 374, 894–897 (2021).
Mallinson, S. J. et al. A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion. Nat. Commun. 9, 2487 (2018).
Sessions, A. L. & Hayes, J. M. Calculation of hydrogen isotopic fractionations in biogeochemical systems. Geochim. Cosmochim. Acta 69, 593–597 (2005).
Sturt, H. F., Summons, R. E., Smith, K., Elvert, M. & Hinrichs, K. U. Intact polar membrane lipids in prokaryotes and sediments deciphered by high‐performance liquid chromatography/electrospray ionization multistage mass spectrometry—new biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrom. 18, 617–628 (2004).
Zhu, C. et al. Comprehensive glycerol ether lipid fingerprints through a novel reversed phase liquid chromatography–mass spectrometry protocol. Org. Geochem. 65, 53–62 (2013).
Huguet, C. et al. An improved method to determine the absolute abundance of glycerol dibiphytanyl glycerol tetraether lipids. Org. Geochem. 37, 1036–1041 (2006).
Becker, K. W., Lipp, J. S., Zhu, C., Liu, X.-L. & Hinrichs, K.-U. An improved method for the analysis of archaeal and bacterial ether core lipids. Org. Geochem. 61, 34–44 (2013).
Pancost, R. D. et al. Archaeol as a methanogen biomarker in ombrotrophic bogs. Org. Geochem. 42, 1279–1287 (2011).
Zhou, Z., Pan, J., Wang, F., Gu, J.-D. & Li, M. Bathyarchaeota: globally distributed metabolic generalists in anoxic environments. FEMS Microbiol. Rev. 42, 639–655 (2018).
Jorgensen, S. L. et al. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc. Natl Acad. Sci. USA 109, E2846–E2855 (2012).
Yu, T., Liang, Q., Niu, M. & Wang, F. High occurrence of Bathyarchaeota (MCG) in the deep‐sea sediments of South China Sea quantified using newly designed PCR primers. Environ. Microbiol. Rep. 9, 374–382 (2017).
Stoddard, S. F., Smith, B. J., Hein, R., Roller, B. R. & Schmidt, T. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 43, D593–D598 (2015).
Hoshino, T. & Inagaki, F. Abundance and distribution of Archaea in the subseafloor sedimentary biosphere. ISME J. 13, 227–231 (2019).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Dong, L. et al. Mass spectrometry data for archaeal ether lipid identification in marine Bathyarchaeia. Zenodo https://doi.org/10.5281/zenodo.16742269 (2025).
Dong, L. et al. Stable carbon isotope data from 13C-labelling experiments tracing carbon assimilation in marine Bathyarchaeia. Zenodo https://doi.org/10.5281/zenodo.16748030 (2025).
Boron, W. F. Evaluating the role of carbonic anhydrases in the transport of HCO3−-related species. Biochim. Biophys. Acta. 1804, 410–421 (2010).
Acknowledgements
We thank W. Wu for his insightful suggestions on stable isotope probing techniques, as well as L. Deng, S. Ding, H. Zhang and H. Hu for their valuable comments during the revision process. We also thank W. Zhang and Q. Luo at the State Key Laboratory of Microbial Metabolism, School of Life Science and Biotechnology, Shanghai Jiao Tong University, for their generous support with mass spectrometry analyses. This work was supported financially by the Natural Science Foundation of China (grants 42230401 to F.W. and 42472370 to L.D.), the Postdoctoral Fellowship Program (Grade B) of China Postdoctoral Science Foundation (GZB20240430 to J.H.), 2030 Project of Shanghai Jiao Tong University (grant WH510244001), Global Subseafloor Ecosystem and Sustainability (GSES) and the project ‘Ocean Negative Carbon Emission (ONCE)’.
Author information
Authors and Affiliations
Contributions
L.D. and F.W. designed the study, and K.-U.H. supervised the study thoroughly. L.D., Y.J., J.H., J.Z., T.Y., S.C., L.L., P.Z. and X.Z. performed the research. L.D., Y.J., J.H., J.Z., S.C., X.Z. and K.-U.H. analysed the data. L.D., Y.J., K.-U.H. and F.W. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Vincent Grossi, Bernhard Naafs and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Quantitative analysis of cellular lipid content in cultured samples.
Quantification of cellular core lipids content in isoprenoid glycerol dibiphytanyl glycerol tetraethers (IPL-derived CLs) following acid hydrolysis over a harvest time of 120 days. The dashed lines indicate the lower and upper bounds (0.7 and 1.5 fg/cell, respectively) for the estimated cellular lipid content derived from a cellular geometry model. Bathyarchaeia biomass was additionally quantified using quantitative PCR (qPCR).
Extended Data Fig. 2 Extracted mass chromatogram of core lipids.
The core lipids were derived from normal-phase UPLC-APCI-QTOF analysis of extract obtained from acid hydrolysis of Bathyarchaeia cells.
Extended Data Fig. 3 Archaeal community structure and core lipid composition following cell acid hydrolysis.
a, Abundance of Bathyarchaeia, estimated by 16S rRNA gene copy numbers after 30 days’ incubation. Data are presented as mean values ± SD (n = 3 biological replicates; independent culture bottles). b, Archaeal community composition at the phylum/class level (left) and Bathyarchaeia subgroup/genus level (right). The Bathyarchaeia community was primarily composed of Baizosediminiarchaeum (93.4%), with Baizomonas comprising 6.5%. “Others” refers to unclassified archaeal taxa. c, Composition of core lipids (CLs) released after acid hydrolysis of archaeal cells. The left bar shows the total distribution of major lipid classes—archaeol (AR), butanetriol dialkyl glycerol tetraethers (BDGTs), and glycerol dialkyl glycerol tetraethers (GDGTs). The right bar displays the relative proportions of individual BDGT (BDGT-0 to BDGT-2) and GDGT (GDGT-0 to GDGT-3) homologues.
Extended Data Fig. 4 Proposed carbon isotope fractionation and metabolic pathways in Bathyarchaeia.
This schematic highlight key carbon isotope fractionation processes and metabolic pathways involved in lipids synthesis by Bathyarchaeia. δ¹³C values and fractionation factors (ε) for different carbon pools and metabolites are shown. The primary carbon sources, dissolved inorganic carbon (DIC) and lignin, are utilization at an approximate ratio of 1:2. Fractionation factors between BP0 and DIC (εBP0/DIC), and BP0 and lignin (εBP0/Lignin) are -36.1‰ and -52.1‰, respectively. The pathways illustrated include demethylation (conversion of lignin to -CH₃-THMPT), acetyl-CoA formation (from acetate), the Wood-Ljungdahl (WL) pathway (conversion of CO₂ to acetyl-CoA via carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS))11, and the role of carbonic anhydrase (CA) in HCO₃⁻ to CO₂ conversion65. This schematic emphasizes the contributions of DIC and lignin to lipid biosynthesis and highlights the isotope fractionation associated with these metabolic pathways.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dong, L., Jing, Y., Hou, J. et al. A dominant subgroup of marine Bathyarchaeia assimilates organic and inorganic carbon into unconventional membrane lipids. Nat Microbiol 10, 2579–2590 (2025). https://doi.org/10.1038/s41564-025-02121-5
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41564-025-02121-5