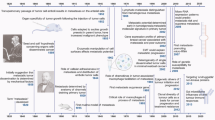
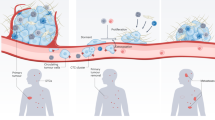
Abstract
The evolution of metastasis in humans is considerably less well understood than the biology of early carcinogenesis. For over a century, clinicians and scientists have been debating whether metastatic potential is the intrinsic property of a cancer, pre-determined by the molecular characteristics of the tumour founder cell, or whether metastatic capacity evolves in a stepwise fashion as the tumour grows, akin to the multistage accumulation of oncogenic alterations that give rise to the first cancer cell. In this Perspective, I examine how genetic analyses of primary tumours and matched metastases can distinguish between these two competing metastasis evolution models, with particular emphasis on the utility of metastatic randomness — a quantitative measure that reflects whether metastases arise from a random selection of primary tumour subclones or whether they are enriched for descendants of privileged lineages that have acquired pro-metastatic traits. Probable metastasis evolution trajectories in tumours with high and low baseline metastatic capacity are discussed, along with the role of seeding rates and selection at different metastatic host sites. Finally, I argue that trailblazing insights into human metastasis biology are immediately possible if we make a concerted effort to apply existing experimental and theoretical tools to the right patient cohorts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Figure 2a shows the replotting of data previously published by Gundem et al.36. Subclone fractions across different lesions are taken directly from Supplementary Table 3. Figure 2b has been redrawn from Figure S14 from Reiter et al.12. Figure 5c,d,e have been redrawn from Fig. 2a,e,h, respectively from Reiter et al.38.
References
Armitage, P. & Doll, R. The age distribution of cancer and a multi-stage theory of carcinogenesis. Br. J. Cancer 8, 1–12 (1954).
Armitage, P. & Doll, R. A two-stage theory of carcinogenesis in relation to the age distribution of human cancer. Br. J. Cancer 11, 161–169 (1957).
Klein, G. Foulds’ dangerous idea revisited: the multistep development of tumors 40 years later. Adv. Cancer Res. 72, 1–23 (1998).
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990).
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013).
Cairns, J. Mutation selection and the natural history of cancer. Nature 255, 197–200 (1975).
Kakiuchi, N. & Ogawa, S. Clonal expansion in non-cancer tissues. Nat. Rev. Cancer 21, 239–256 (2021).
Nowell, P. C. The clonal evolution of tumor cell populations: acquired genetic lability permits stepwise selection of variant sublines and underlies tumor progression. Science 194, 23–28 (1976).
Priestley, P. et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 575, 210–216 (2019).
Mlecnik, B. et al. The tumor microenvironment and Immunoscore are critical determinants of dissemination to distant metastasis. Sci. Transl. Med. 8, 327ra26 (2016).
Nguyen, B. et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell 185, 563–575.e11 (2022).
Reiter, J. G. et al. Minimal functional driver gene heterogeneity among untreated metastases. Science 361, 1033–1037 (2018).
Makohon-Moore, A. P. et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat. Genet. 49, 358–366 (2017).
Reiter, J. G. et al. An analysis of genetic heterogeneity in untreated cancers. Nat. Rev. Cancer 19, 639–650 (2019).
Fisher, B. Laboratory and clinical research in breast cancer — a personal adventure: the David A. Karnofsky memorial lecture. Cancer Res. 40, 3863–3874 (1980).
Fisher, B. et al. Twenty-five-year follow-up of a randomized trial comparing radical mastectomy, total mastectomy, and total mastectomy followed by irradiation. N. Engl. J. Med. 347, 567–575 (2002).
Klein, C. A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer 9, 302–312 (2009).
Hüsemann, Y. et al. Systemic spread is an early step in breast cancer. Cancer Cell 13, 58–68 (2008).
Hosseini, H. et al. Early dissemination seeds metastasis in breast cancer. Nature 540, 552–558 (2016).
Weinberg, R. A. Mechanisms of malignant progression. Carcinogenesis 29, 1092–1095 (2008).
Fidler, I. J. & Kripke, M. L. Metastasis results from preexisting variant cells within a malignant tumor. Science 197, 893–895 (1977).
Minn, A. J. et al. Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 (2005).
Kang, Y. et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537–549 (2003).
Reticker-Flynn, N. E. et al. Lymph node colonization induces tumor-immune tolerance to promote distant metastasis. Cell 185, 1924–1942.e23 (2022).
Sottoriva, A. et al. A Big Bang model of human colorectal tumor growth. Nat. Genet. 47, 209–216 (2015).
Watkins, T. B. K. et al. Pervasive chromosomal instability and karyotype order in tumour evolution. Nature 587, 126–132 (2020).
Heide, T. et al. The co-evolution of the genome and epigenome in colorectal cancer. Nature 611, 733–743 (2022).
Moorman, A. et al. Progressive plasticity during colorectal cancer metastasis. Nature 637, 947–954 (2025).
Halsted, W. S. I. The results of operations for the cure of cancer of the breast performed at the Johns Hopkins hospital from June, 1889, to January, 1894. Ann. Surg. 20, 497–555 (1894).
Hellman, S. Karnofsky memorial lecture. Natural history of small breast cancers. J. Clin. Oncol. 12, 2229–2234 (1994).
Jamal-Hanjani, M. et al. Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121 (2017).
Zhao, Y. et al. Selection of metastasis competent subclones in the tumour interior. Nat. Ecol. Evol. 5, 1033–1045 (2021).
Mathur, R. et al. Glioblastoma evolution and heterogeneity from a 3D whole-tumor perspective. Cell 187, 446–463.e16 (2024).
Cross, W. et al. The evolutionary landscape of colorectal tumorigenesis. Nat. Ecol. Evol. 2, 1661–1672 (2018).
Ryser, M. D., Min, B.-H., Siegmund, K. D. & Shibata, D. Spatial mutation patterns as markers of early colorectal tumor cell mobility. Proc. Natl Acad. Sci. USA 115, 5774–5779 (2018).
Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Reiter, J. G. et al. Lymph node metastases develop through a wider evolutionary bottleneck than distant metastases. Nat. Genet. 52, 692–700 (2020).
Hu, Z., Li, Z., Ma, Z. & Curtis, C. Multi-cancer analysis of clonality and the timing of systemic spread in paired primary tumors and metastases. Nat. Genet. 52, 701–708 (2020).
Cheung, K. J. & Ewald, A. J. A collective route to metastasis: seeding by tumor cell clusters. Science 352, 167–169 (2016).
Heyde, A., Reiter, J. G., Naxerova, K. & Nowak, M. A. Consecutive seeding and transfer of genetic diversity in metastasis. Proc. Natl Acad. Sci. USA 116, 14129–14137 (2019).
Aceto, N. et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 (2014).
Abbosh, C. et al. Tracking early lung cancer metastatic dissemination in TRACERx using ctDNA. Nature 616, 553–562 (2023).
Aldea, M. et al. Overcoming resistance to tumor-targeted and immune-targeted therapies. Cancer Discov. 11, 874–899 (2021).
Wassenaar, E. C. E. et al. A unique interplay of access and selection shapes peritoneal metastasis evolution in colorectal cancer. Preprint at bioRxiv https://doi.org/10.1101/2024.09.25.614736 (2024).
Tarabichi, M. et al. A practical guide to cancer subclonal reconstruction from DNA sequencing. Nat. Methods 18, 144–155 (2021).
Bozic, I. et al. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl Acad. Sci. USA 107, 18545–18550 (2010).
Yates, L. R. et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell 32, 169–184.e7 (2017).
Gibson, W. J. et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat. Genet. 48, 848–855 (2016).
Hu, Z. et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat. Genet. 51, 1113–1122 (2019).
Al Bakir, M. et al. The evolution of non-small cell lung cancer metastases in TRACERx. Nature 616, 534–542 (2023).
Brastianos, P. K. et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 5, 1164–1177 (2015).
Naxerova, K. et al. Hypermutable DNA chronicles the evolution of human colon cancer. Proc. Natl Acad. Sci. USA 111, E1889–E1898 (2014).
Naxerova, K. et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science 357, 55–60 (2017).
Blohmer, M. et al. Quantifying cell divisions along evolutionary lineages in cancer. Nat. Genet. 57, 706–717 (2025).
Haffner, M. C. et al. Tracking the clonal origin of lethal prostate cancer. J. Clin. Invest. 123, 4918–4922 (2013).
Liu, W. et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat. Med. 15, 559–565 (2009).
American Cancer Society. Survival rates for pancreatic cancer. https://www.cancer.org/cancer/types/pancreatic-cancer/detection-diagnosis-staging/survival-rates.html (2025).
American Cancer Society. Thyroid cancer survival rates, by type and stage. https://www.cancer.org/cancer/types/thyroid-cancer/detection-diagnosis-staging/survival-rates.html (2024).
LiVolsi, V. A. Papillary thyroid carcinoma: an update. Mod. Pathol. 24, S1–S9 (2011).
Li, Y., Tang, C.-G., Zhao, Y., Cao, W.-Y. & Qu, G.-F. Outcomes and prognostic factors of patients with stage IB and IIA pancreatic cancer according to the 8th edition American Joint Committee on Cancer criteria. World J. Gastroenterol. 23, 2757–2762 (2017).
Baggiolini, A. et al. Developmental chromatin programs determine oncogenic competence in melanoma. Science 373, eabc1048 (2021).
Dongre, A. & Weinberg, R. A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84 (2019).
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010).
Jones, S. et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl Acad. Sci. USA 105, 4283–4288 (2008).
Williams, N. et al. Life histories of myeloproliferative neoplasms inferred from phylogenies. Nature 602, 162–168 (2022).
Sun, Y. et al. Integrated multi-omics profiling to dissect the spatiotemporal evolution of metastatic hepatocellular carcinoma. Cancer Cell 42, 135–156.e17 (2024).
Williams, M. J., Werner, B., Barnes, C. P., Graham, T. A. & Sottoriva, A. Identification of neutral tumor evolution across cancer types. Nat. Genet. 48, 238–244 (2016).
Frankell, A. M. et al. The evolution of lung cancer and impact of subclonal selection in TRACERx. Nature 616, 525–533 (2023).
Martínez-Jiménez, F. et al. Pan-cancer whole-genome comparison of primary and metastatic solid tumours. Nature 618, 333–341 (2023).
Blackford, A. L., Canto, M. I., Klein, A. P., Hruban, R. H. & Goggins, M. Recent trends in the incidence and survival of stage 1A pancreatic cancer: a surveillance, epidemiology, and end results analysis. J. Natl Cancer Inst. 112, 1162–1169 (2020).
Yavuz, A., Meyer, J. J. & Marks, L. B. Association between the rates of synchronous and metachronous metastases: analysis of SEER data. Cancer Network https://www.cancernetwork.com/view/association-between-rates-synchronous-and-metachronous-metastases-analysis-seer-data (2007).
Hong, J. et al. Convergence for inactivation of TGFβ signaling is a common feature of advanced pancreatic cancer. Preprint at medRxiv https://doi.org/10.1101/2024.01.30.24301554 (2024).
Siegel, D. A., O’Neil, M. E., Richards, T. B., Dowling, N. F. & Weir, H. K. Prostate cancer incidence and survival, by stage and race/ethnicity — United States, 2001–2017. MMWR Morb. Mortal. Wkly. Rep. 69, 1473–1480 (2020).
American Cancer Society. Prostate cancer stages. https://www.cancer.org/cancer/types/prostate-cancer/detection-diagnosis-staging/staging.html (2023).
Riihimäki, M., Hemminki, A., Sundquist, J. & Hemminki, K. Patterns of metastasis in colon and rectal cancer. Sci. Rep. 6, 29765 (2016).
Chen, M.-T. et al. Comparison of patterns and prognosis among distant metastatic breast cancer patients by age groups: a SEER population-based analysis. Sci. Rep. 7, 9254 (2017).
Paget, S. The distribution of secondary growths in cancer of the breast. Lancet 133, 571–573 (1889).
Ewing, J. in Neoplastic Diseases (W. B. Saunders, Philadelphia, 1928).
Ildiz, E. S., Gvozdenovic, A., Kovacs, W. J. & Aceto, N. Travelling under pressure — hypoxia and shear stress in the metastatic journey. Clin. Exp. Metastasis 40, 375–394 (2023).
Riihimäki, M., Thomsen, H., Sundquist, K., Sundquist, J. & Hemminki, K. Clinical landscape of cancer metastases. Cancer Med. 7, 5534 (2018).
Shih, D. J. H. et al. Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat. Genet. 52, 371–377 (2020).
Ulintz, P. J., Greenson, J. K., Wu, R., Fearon, E. R. & Hardiman, K. M. Lymph node metastases in colon cancer are polyclonal. Clin. Cancer Res. 24, 2214–2224 (2018).
Sleeman, J. P., Cady, B. & Pantel, K. The connectivity of lymphogenous and hematogenous tumor cell dissemination: biological insights and clinical implications. Clin. Exp. Metastasis 29, 737–746 (2012).
Sleeman, J. P. in Recent Results in Cancer Research Vol. 157 (eds Schlag, P.M. & Veronesi, U.) 55–81 (Springer Berlin Heidelberg, 2000).
Ubellacker, J. M. et al. Lymph protects metastasizing melanoma cells from ferroptosis. Nature 585, 113–118 (2020).
Sleeman, J., Schmid, A. & Thiele, W. Tumor lymphatics. Semin. Cancer Biol. 19, 285–297 (2009).
Väyrynen, V. et al. Incidence and management of patients with colorectal cancer and synchronous and metachronous colorectal metastases: a population-based study. BJS Open. 4, 685–692 (2020).
Küçükköse, E. et al. Lymphatic invasion of plakoglobin-dependent tumor cell clusters drives formation of polyclonal lung metastases in colon cancer. Gastroenterology 165, 429–444.e15 (2023).
American Cancer Society. Colorectal cancer survival rates. https://www.cancer.org/cancer/types/colon-rectal-cancer/detection-diagnosis-staging/survival-rates.html (2025).
Franko, J. et al. Prognosis of patients with peritoneal metastatic colorectal cancer given systemic therapy: an analysis of individual patient data from prospective randomised trials from the Analysis and Research in Cancers of the Digestive System (ARCAD) database. Lancet Oncol. 17, 1709–1719 (2016).
Weichselbaum, R. R. & Hellman, S. Oligometastases revisited. Nat. Rev. Clin. Oncol. 8, 378–382 (2011).
Gallinger, S. et al. Liver resection for colorectal cancer metastases. Curr. Oncol. 20, e255–e265 (2013).
Chen, H.-N. et al. Genomic evolution and diverse models of systemic metastases in colorectal cancer. Gut 71, 322–332 (2022).
Agudo, J. et al. Targeting cancer cell dormancy. Nat. Rev. Cancer 24, 97–104 (2024).
Phan, T. G. & Croucher, P. I. The dormant cancer cell life cycle. Nat. Rev. Cancer 20, 398–411 (2020).
Dai, J. et al. Astrocytic laminin-211 drives disseminated breast tumor cell dormancy in brain. Nat. Cancer 3, 25–42 (2022).
Zhang, C. et al. Mapping the spreading routes of lymphatic metastases in human colorectal cancer. Nat. Commun. 11, 1–11 (2020).
Venet, D. et al. Phylogenetic reconstruction of breast cancer reveals two routes of metastatic dissemination associated with distinct clinical outcome. eBioMedicine 56, 102793 (2020).
Orrantia-Borunda, E., Anchondo-Nuñez, P., Acuña-Aguilar, L. E., Gómez-Valles, F. O. & Ramírez-Valdespino, C. A. in Breast Cancer (ed. Mayrovitz, H. N.) 31–42 (Exon Publications, 2022).
Hebert, J. D., Neal, J. W. & Winslow, M. M. Dissecting metastasis using preclinical models and methods. Nat. Rev. Cancer 23, 391–407 (2023).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Zack, T. I. et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013).
Turajlic, S. et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell 173, 581–594.e12 (2018).
American Cancer Society. Survival rates for prostate cancer. https://www.cancer.org/cancer/types/prostate-cancer/detection-diagnosis-staging/survival-rates.html (2025).
Boire, A. et al. Why do patients with cancer die? Nat. Rev. Cancer 24, 578–589 (2024).
Acknowledgements
I thank members of the Naxerova lab, M. Nicholson and X. Brunet-Guasch for helpful discussions and for critical reading of the manuscript. This work was supported by the National Cancer Institute of the US National Institutes of Health (R37CA225655, R01CA279054, R01CA26928) and by an Emerging Leader Award from the Mark Foundation for Cancer Research.
Author information
Authors and Affiliations
Contributions
The author contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Peer review
Peer review information
Nature Reviews Cancer thanks Alvin Makohon-Moore, Nathan Reticker-Flynn who co-reviewed with Marcos Labrado and the other, anonymous, reviewer for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Epithelial-to-mesenchymal transition
-
The process by which cells lose epithelial characteristics (e.g. apicobasal polarity, cell–cell adhesions) and acquire mesenchymal properties (increased motility and invasiveness).
- Ferroptosis
-
Iron-dependent form of cell death.
- Genetic drift
-
The change of allele frequencies in a population over time due to chance alone.
- Metachronously
-
Referring to metastases that are discovered more than 3–6 months after the primary tumour diagnosis.
- Microsatellite
-
DNA sequences composed of short repeats. Microsatellites mutate at higher rates than most other genomic regions.
- Peritoneum
-
The lining of the abdominal cavity which holds in place and protects most abdominal organs, including the intestines.
- Synchronously
-
Referring to metastases that are diagnosed at the same time as the primary tumour.
- Variant allele frequencies
-
Fractions of variant (non-germline) alleles in a tissue sample. The variant allele frequency reflects the proportion of mutant cells in a population.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Naxerova, K. Evolutionary paths towards metastasis. Nat Rev Cancer 25, 545–560 (2025). https://doi.org/10.1038/s41568-025-00814-x
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41568-025-00814-x
This article is cited by
-
Genomic evolution of pancreatic cancer at single-cell resolution
Nature Genetics (2026)