Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the cause of coronavirus disease 2019 (COVID-19), which has become a public health emergency of international concern1. Angiotensin-converting enzyme 2 (ACE2) is the cell-entry receptor for severe acute respiratory syndrome coronavirus (SARS-CoV)2. Here we infected transgenic mice that express human ACE2 (hereafter, hACE2 mice) with SARS-CoV-2 and studied the pathogenicity of the virus. We observed weight loss as well as virus replication in the lungs of hACE2 mice infected with SARS-CoV-2. The typical histopathology was interstitial pneumonia with infiltration of considerable numbers of macrophages and lymphocytes into the alveolar interstitium, and the accumulation of macrophages in alveolar cavities. We observed viral antigens in bronchial epithelial cells, macrophages and alveolar epithelia. These phenomena were not found in wild-type mice infected with SARS-CoV-2. Notably, we have confirmed the pathogenicity of SARS-CoV-2 in hACE2 mice. This mouse model of SARS-CoV-2 infection will be valuable for evaluating antiviral therapeutic agents and vaccines, as well as understanding the pathogenesis of COVID-19.
Similar content being viewed by others
Main
In late December 2019, cases of COVID-19—which is caused by SARS-CoV-2—were identified and reported from Wuhan city (Hubei province, China), where they were linked to a seafood market at which exotic animals were also sold and consumed1,3. The number of confirmed cases has since soared: as of 25 February 2020, almost 78,000 cases and over 2,700 deaths were reported in China4, and imported cases from travellers from mainland China were reported in several other countries. It is critical to establish the pathogenicity and biology of the virus for prevention and treatment of the disease.
Because SARS-CoV-2 is highly homologous with SARS-CoV, human ACE2—which is the entry receptor of SARS-CoV—was also considered to have a high binding ability with the SARS-CoV-2 by molecular biological analysis2,5. We therefore used transgenic hACE2 mice and wild-type mice infected with the HB-01 strain of SARS-CoV-2 to study the pathogenicity of the virus.
Specific-pathogen-free male and female wild-type (n = 15) or hACE2 (n = 19) mice of 6–11 months of age were inoculated intranasally with SARS-CoV-2 strain HB-01 at a dosage of 105 50% tissue culture infectious dose (TCID50) per 50 μl inoculum volume per mouse, after the mice were intraperitoneally anaesthetized using 2.5% avertin; mock-treated hACE2 mice (n = 15) were used as control. Clinical manifestations were recorded from 13 mice (3 HB-01-infected wild-type mice; 3 mock-treated hACE2 mice; and 7 HB-01-infected hACE2 mice). We observed slight bristled fur and weight loss only in the HB-01-infected hACE2 mice—and not the HB-01-infected wild-type mice or mock-treated hACE2 mice—during the 14 days of observation; other clinical symptoms, such as an arched back and decreased response of external stimuli, were not found in any of the mice. Notably, the weight loss of HB-01-infected hACE2 mice was up to 8% at 5 days post-infection (dpi) (Fig. 1a).
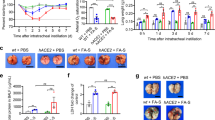
a, Weight loss was recorded for 14 days. hACE2 mice (n = 7) and wild-type (WT) mice (n = 3) were experimentally challenged intranasally with SARS-CoV-2 HB-01, and mock-treated hACE2 (ACE2 + mock) mice (n = 3) were used as control. According to two-tailed Mann–Whitney U-test, the weight of HB-01-infected hACE2 mice (ACE2 + HB-01) displayed a significant decline compared with that of HB-01-infected wild-type mice (WT + HB-01) or mock-treated hACE2 mice (***P = 0.0005). b, To measure viral RNA, 12 mice were infected in each group. Three mice per group were killed, and their major organs (including testis in male mice) were collected for analysis of viral loads and virus titre at 1, 3, 5 and 7 dpi. The distribution of SARS-CoV-2 in the primary organs of HB-01-infected hACE2 mice was detected using RT–qPCR. c, Virus titres in the lungs were determined on Vero E6 cells. According to a two-tailed unpaired Welch’s t-test, viral titres in the lungs from HB-01-infected hACE2 mice (n = 3) showed a significant increase compared with those in HB-01-infected wild-type mice (n = 3) or mock-treated hACE2 mice (n = 3) at 1 (**P = 0.0053), 3 (**P = 0.0022) and 5 (**P = 0.0081) dpi. d, Virus isolated from the lungs of HB-01-infected hACE2 mice at 3 dpi was observed using electron microscopy. Scale bar, 200 nm. Data are representative of three independent experiments. e, The specific IgG against SARS-CoV-2 was detected from the sera of mice (HB-01-infected wild-type (n = 3) or hACE2 ( n =7) mice) at day 0 and 21 dpi by enzyme-linked immunosorbent assay (ELISA). OD450, optical density at 450 nm. Two-tailed unpaired Student’s t-test; not significant (NS), P = 0.2193; two-tailed unpaired Welch’s t-test, ***P = 3.11 × 10−6. Data in a–c, e are mean ± s.d.
Next, we examined viral replication and pathological changes in three mice per group at each time point; the primary organs—including heart, liver, spleen, lung, kidney, brain, intestine and testis—were collected periodically. As shown in Fig. 1b, viral loads were detectable by quantitative PCR with reverse transcription (RT–qPCR) at 1, 3, 5 and 7 dpi in the lungs of HB-01-infected hACE2 mice (but not in those of HB-01-infected wild-type mice; data not shown), and viral RNA copies reached a peak of 106.77 copies per ml at 3 dpi. Viral RNA was also detectable at 1 dpi in the intestine of HB-01-infected hACE2 mice, which was not detected in other tissues along the timeline (Fig. 1b). Although viral loads were detectable in the intestine, no virus in the intestine was isolated at 1 dpi; we therefore speculate that the viral load detected was residual input inoculum from the nasal mucosa transferred to the intestines by swallowing. Consistent with the results showing viral loads in the lung, infectious virus was isolated from the lungs of HB-01-infected hACE2 mice at 1, 3 and 5 dpi; the highest virus titres were detected at 3 dpi (102.44 TCID50 per 100 μl) (Fig. 1c). We isolated infectious virus using Vero E6 cell culture from the lung, and observed SARS-CoV-2 particles using electron microscopy (Fig. 1d). However, the virus was not isolated from the lungs of HB-01-infected wild-type mice or mock-treated hACE2 mice along the detecting timeline (Fig. 1c), which suggests that human ACE2 is essential for SARS-CoV-2 infection and replication in mice. Moreover, we found specific IgG antibodies against the spike (S) protein of SARS-CoV-2 in the sera of HB-01-infected hACE2 mice at 21 dpi (Fig. 1e).
There were no obviously gross pathological or histopathological changes at 1 dpi in any of the mice. Compared with HB-01-infected wild-type mice or mock-treated hACE2 mice (both of which showed homogeneously pink and slightly deflated lung lobes), HB-01-infected hACE2 mice at 3 dpi displayed gross lesions with focal-to-multifocal dark-red discoloration in some of the lung lobes. The lesions progressed into multifocal-to-coalescent scattered dark-reddish-purple areas and focal palpable nodules throughout the lung lobes at 5 dpi (Fig. 2a). The damaged lungs became swollen and enlarged. Microscopically, the lung tissues from HB-01-infected hACE2 mice at 3 dpi displayed moderate interstitial pneumonia, characterized by thickened alveolar septa accompanied by infiltration of inflammatory cells in 70–80% of the lung tissues, and an accumulation of inflammatory cells in partial alveolar cavities (20–30%). Inflammatory cells—including lymphocytes, macrophages and neutrophils—accumulated in the alveolar interstitium and caused thickening of the alveolar walls. At 5 dpi, the lung progressed into coalescing interstitial pneumonia with diffuse lesions, characterized by more-severe thickened alveolar septa accompanied with infiltration of inflammatory cells, and accumulation of inflammatory cells in more alveolar cavities (40–50%). The thickened alveolar septa were filled with macrophages, lymphocytes and neutrophils (Fig. 2a). A small amount of collagen fibre was observed in the thickened alveolar interstitium in the HB-01-infected hACE2 mice by modified Masson’s trichrome stain at 5 dpi (Extended Data Fig. 1a). The bronchiolar epithelial cells showed swelling and degeneration; some of these cells fragmented (Fig. 2a). A few periodic-acid–Schiff-positive-exudation, or denatured and detached, bronchiolar epithelia were occasionally observed in affected bronchioles at 5 dpi (Extended Data Fig. 1b). The alveolar cavities were distended mainly by swollen and degenerative macrophages, lymphocytes and neutrophils (Fig. 2a). To investigate the infiltration of specific inflammatory cells, immunohistochemistry (IHC) was carried out to identify MAC2+ macrophages (Extended Data Fig. 2a), CD3+ T lymphocytes and CD19+ B lymphocytes (Extended Data Fig. 2b). Compared to the lungs of HB-01-infected wild-type mice, more macrophages and T lymphocytes were found in the lungs of HB-01-infected hACE2 mice and their numbers increased along with the prolonged infection time. MAC2+ macrophages were diffusely infiltrated into the alveolar cavities (at 3 dpi) or focally aggregated in the thickened alveolar septa (at 5 dpi) (Extended Data Fig. 2a). CD3+ T lymphocytes were dispersed or (occasionally) aggregated in the alveolar interstitium in HB-01-infected hACE2 mice at 3 and 5 dpi, and some CD19+ B lymphocytes were also observed at 5 dpi (Extended Data Fig. 2b). Perivascular infiltrating inflammatory cells—including lymphocytes, macrophages and neutrophils—were observed multifocally within and adjacent to affected areas of the lungs. In lung lesions, IHC staining of sequential sections revealed that viral antigens were found in macrophages, alveolar epithelia and in bronchial epithelial cells that were degenerative and being desquamated (Fig. 2b). We also observed small amounts of viral antigen in respiratory epithelial cells in areas of the lungs that did not show lesions (data not shown). However, there were no substantial histopathological changes (Extended Data Fig. 3) or viral antigens for SARS-CoV-2 (Extended Data Fig. 4) in the other organs, including myocardium, liver, spleen, kidney, cerebrum, intestine and testis. At 7 dpi, the pneumonia became mild with focal lesion areas (data not shown).
a, Gross pathology and histopathology of lungs from HB-01-infected wild-type mice (3 dpi), mock-treated hACE2 mice (3 dpi) and HB-01-infected hACE2 mice (3 and 5 dpi). Post-mortem examinations showed focal dark-red lesions (red arrow) throughout the dorsal region of the right middle lobe of the lung at 3 dpi. The lesions progressed into multifocally scattered dark-reddish-purple areas and palpable nodules (red arrow) throughout the right lobe of the lung at 5 dpi. Histopathological observations indicated that moderate interstitial pneumonia with thickened alveolar septa (black arrows) and infiltration of lymphocytes (red frames, 1,000× magnification). The swollen and degenerative mononuclear cells (green frames, 1,000× magnification) are scattered within the alveolar cavities at 3 and 5 dpi. b, IHC examination of the lungs of each mouse group. The sequential sections were stained by haematoxylin and eosin (H&E) or IHC. Viral antigens were observed in the mononuclear cells (green arrows), alveolar epithelia (blue arrows) and bronchial epithelial cells that were degenerative and being desquamated (black arrows). The black frames in the top-right corners are a magnification of the region in the respective black box. Scale bars, 100 μm (black), 50 μm (red). Data in a, b are representative of three independent experiments.
In addition, we demonstrated the colocalization of SARS-CoV-2 S protein (Fig. 3f) and the human ACE2 receptor (Fig. 3g) in alveolar epithelial cells of HB-01-infected hACE2 mice using immunofluorescence, at 3 dpi (Fig. 3h). This phenomenon was not found in mock-treated hACE2 mice (Fig. 3a–d) or HB-01-infected wild-type mice (data not shown), which indicates that SARS-CoV-2—as with SARS-CoV—uses the human ACE2 as a receptor for entry5.
Colocalization of SARS-CoV-2 S protein and human ACE2 receptor in the lungs of hACE2 mice. The sections were incubated with anti-SARS-CoV-2 S protein antibody, anti-human ACE2 antibody and DAPI. a–d, Lung sections of mock-treated hACE2 mice. e–h, Lung sections of HB-01-infected hACE2 mice. White arrows show the viral S protein (f) and human ACE2 (g); yellow arrows show the merge of viral S protein and human ACE2 (h). Scale bars, 25 μm. Data in a–h are representative of three independent experiments.
The speed of the geographical spread of COVID-19 has led to the disease being declared a public health emergency of international concern, with cases reported on multiple continents only weeks after the disease was first reported6. Although it has been determined by bioinformatics that the pathogen that causes this epidemic is a novel coronavirus, it is necessary that this is confirmed by animal experiments (following Koch’s postulates)7. Previous clinical studies have confirmed the isolation of the virus from hosts with the disease and cultivation in host cells1. Here we show that, after the experimental infection of hACE2 mice with one of the earliest known isolates of SARS-CoV-2, our mouse model of SARS-CoV-2 infection exhibits viral replication in the lungs characterized by moderate interstitial pneumonia—similar to initial clinical reports of pneumonia caused by SARS-CoV-28. Moreover, we also observed specific antibodies against SARS-CoV-2 and re-isolated the virus from infected mice.
The fatality rate of currently reported cases of COVID-19 is about 2%, which implies that—so far—SARS-CoV-2 does not seem to cause the high fatality rates seen for SARS-CoV (9–11%)9; this suggests that there are differences in pathogenicity between the two viruses. In mice, the pathogenicity of SARS-CoV-2 seems mild compared to SARS-CoV; the latter causes extrapulmonary organ damage (including the brain, kidney, intestine, heart and liver) and, furthermore, neurons are susceptible to infection with SARS-CoV—cerebral vasculitis and haemorrhage were observed in SARS-CoV-infected hACE2 mice10,11. However, only interstitial pneumonia was observed in SARS-CoV-2-infected hACE2 mice, which implies a disparity in pathogenicity between the two coronaviruses.
Our results demonstrate the pathogenicity of SARS-CoV-2 in mice, which—together with previous clinical studies1—completely satisfies Koch’s postulates7 and confirms that SARS-CoV-2 is the pathogen responsible for COVID-19. Our mouse model of SARS-CoV-2 infection will be valuable for evaluating antiviral therapeutic agents and vaccines, as well as understanding the pathogenesis of this disease.Note added in proof: In the version of this paper that was originally published online, Fig. 2a contained a duplication. In the version of the figure that was originally published, tissue sections from a WT + HB-01 sample were shown instead of tissue sections from ACE2 + mock. The originally published figure can be found here as Supplementary Fig. 1.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized and investigators were not blinded to allocation during experiments and outcome assessment.
Ethics statement
Mouse studies were performed in an animal biosafety level 3 (ABSL3) facility using HEPA-filtered isolators. All procedures in this study involving mice were reviewed and approved by the Institutional Animal Care and Use Committee of the Institute of Laboratory Animal Science, Peking Union Medical College (BLL20001). All of the experiments complied with all relevant ethical regulations.
Viruses and cells
The SARS-CoV-2 strain HB-01 was provided by W. Tan1. The complete genome for this SARS-CoV-2 has been submitted to GISAID (identifier: BetaCoV/Wuhan/IVDC-HB-01/2020|EPI_ISL_402119), and deposited in the China National Microbiological Data Center (accession number NMDC10013001 and genome accession number MDC60013002-01). Seed SARS-CoV-2 stocks and virus isolation studies were performed in Vero cells, which are maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin, and 100 μg/ml streptomycin, and incubated at 37 °C, 5% CO2. For infected mice, lung homogenates were used for virus titration tests using endpoint titration in Vero E6 cells. Virus titres of the supernatant were determined using a standard TCID50 assay.
Mouse experiments
For the mouse experiments, specific-pathogen-free, 6–11-month-old male and female hACE2 mice were obtained from the Institute of Laboratory Animal Science, Peking Union Medical College. Transgenic mice were generated by microinjection of the mouse Ace2 promoter driving the human ACE2 coding sequence into the pronuclei of fertilized ova from ICR mice, and then human ACE2 integrated was identified by PCR as previous described10; the human ACE2 mainly expressed in the lungs, heart, kidneys and intestines of transgenic mice. After being intraperitoneally anaesthetized by 2.5% avertin with 0.02 ml/g body weight, the hACE2 or wild-type (ICR) mice were inoculated intranasally with SARS-CoV-2 stock virus at a dosage of 105 TCID50, and hACE2 mice intranasally inoculated with an equal volume of PBS were used as a mock-infection control. The infected mice were continuously observed to record body weight, clinical symptoms, responses to external stimuli and death. Mice were dissected at 1, 3, 5 and 7 dpi to collect different tissues to screen virus replication and histopathological changes.
Preparation of homogenate supernatant
Tissues homogenates (1 g/ml) were prepared by homogenizing perfused tissues using an electric homogenizer for 2 min 30 s in DMEM. The homogenates were centrifuged at 3,000 rpm for 10 min at 4 °C. The supernatant was collected and stored at −80 °C for viral titre and viral load detection.
RNA extraction and RT–qPCR
Total RNA was extracted from tissues homogenates of organs using the RNeasy Mini Kit (Qiagen), and reverse transcription was performed using the PrimerScript RT Reagent Kit (TaKaRa) following the manufacturers’ instructions. RT–qPCR reactions were performed using the PowerUp SYBG Green Master Mix Kit (Applied Biosystems), in which samples were processed in duplicate using the following cycling protocol: 50 °C for 2 min, 95 °C for 2 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 30 s, and then 95 °C for 15 s, 60 °C for 1 min, 95 °C for 45 s. The primer sequences used for RT–qPCR are targeted against the envelope (E) gene of SARS-CoV-2 and are as follows: forward: 5′-TCGTTTCGGAAGAGACAGGT-3′, reverse: 5′-GCGCAGTAAGGATGGCTAGT-3′. The PCR products were verified by sequencing using the dideoxy method on an ABI 3730 DNA sequencer (Applied Biosystems). During the sequencing process, amplification was performed using specific primers. The sequences for this process are available upon request to the corresponding author. The sequencing reads obtained were linked using DNAMAN, and the results were compared using the Megalign module in the DNAStar software package. The SYBR green real-time PCR standard curve was generated by serial tenfold dilutions of recombinant plasmid with a known copy number (from 1.47 × 109 to 1.47 × 101 copies per μl). These dilutions were tested and used as quantification standards to construct the standard curve by plotting the plasmid copy number against the corresponding threshold cycle values (Ct). Results were expressed as log10-transformed numbers of genome equivalent copies per ml of sample.
ELISA method
The specific IgG against SARS-CoV-2 from HB-01-infected hACE2 and wild-type mice was determined by ELISA. Ninety-six-well plates were coated with the Spike 1 (S1) protein of SARS-CoV-2 (0.1 μg/100 μl, Sino Biological, 40591-V08H), the tested sera were diluted at 1:100 and added to each well, and 3 multiple wells were set for each sample, and then incubated at 37 °C for 30 min, followed by goat anti-mouse secondary antibodies conjugated with HRP (ZB-2305, zhongshan,1:10,000 dilution), and incubated at room temperature for 30 min. The reaction was developed by TMB substrate and the optical densities at 450 nm were determined (Metertech960 enzyme marker with 450 nm wavelength).
Laboratory preparation of the antibody of SARS-CoV-2 S1 protein
Mice were immunized with purified SARS-CoV-2 S1 protein (Sino biological) and splenocytes of hyper-immunized mice were fused with myeloma cells. Positive clones were selected by ELISA using SARS-CoV-2 S1 protein (Extended Data Fig. 5). The cell supernatant of 7D2 clone, which binds to the SARS-CoV-2 S1 protein, was collected for immunofluorescence analysis.
Pathological examination
Autopsies were performed in an animal biosafety level 3 (ABSL3) laboratory. Major organs were grossly examined and then fixed in 10% buffered formalin solution, and paraffin sections (3–4 μm in thickness) were prepared routinely. Haematoxylin and eosin, periodic acid–Schiff and modified Masson’s trichrome stains were used to identify histopathological changes in all of the organs. The histopathology of the lung tissue was observed by light microscopy.
IHC
The organs were fixed in 10% buffered formalin solution, and paraffin sections (3–4 μm in thickness) were prepared routinely. Sections were treated with an antigen retrieval kit (Boster, AR0022) for 1 min at 37 °C and quenched for endogenous peroxidases in 3% H2O2 in methanol for 10 min. After blocking in 1% normal goat serum, the sections were incubated with 7D2 monoclonal antibody (laboratory preparation) at 4 °C overnight, followed by HRP-labelled goat anti-mouse IgG secondary antibody (Beijing ZSGB Biotechnology, ZDR-5307). Alternatively, the sections were stained with rat IgG2a antibody (Abcam, ab18450, RTK2758), MAC2 antibody (Cedarlane Laboratories, CL8942AP), CD3 antibody (Dako, A0452) or CD19 antibody (Cell Signaling Technology, 3574) at 4 °C overnight. Subsequently, the sections were incubated with goat anti-rat IgG secondary antibody (HRP) (Beijing ZSGB Biotechnology, PV9004) or goat anti-rabbit IgG secondary antibody (HRP) (Beijing ZSGB Biotechnology, PV9001) for 60 min, and visualized by 3,30-diaminobenzidine tetrahydrochloride (DAB). The slices were counterstained with haematoxylin, dehydrated and mounted on a slide and viewed under an Olympus microscope. The sections from HB-01-infected wild-type mice, mock-treated hACE2 mice or HB-01-infected hACE2 mice were directly incubated with HRP-labelled goat anti-rat/mouse/rabbit IgG and used as the negative control for MAC2, CD19, CD3 or viral antigen staining. Rat IgG2a antibody was used as isotype control for MAC2 staining. For the expression of viral antigen, the sections from HB-01-infected wild-type mice or mock-treated hACE2 mice incubated with anti-S protein antibody was also used as a negative control.
Confocal microscopy
For analysis of the colocalization of viruses and the human ACE2 receptor, the lung tissue sections were washed twice with PBS, fixed by Immunol staining fix solution (P0098, Beyotime Biotechnology), blocked for 1 h at room temperature by Immunol staining blocking buffer (P0102, Beyotime Biotechnology) and then incubated overnight at 4 °C with the appropriate primary and secondary antibodies. The nuclei were stained with DAPI. Anti-S protein antibody (mouse monoclonal 7D2, laboratory preparation, 1:200) and anti-hACE2 antibody (rabbit polyclonal, ab15348, Abcam, 1:200) were used as the primary antibodies. The sections were washed with PBS and incubated with secondary antibodies conjugated with FITC (goat anti-mouse, ZF-0312, Beijing ZSGB Biotechnology, 1:200) and TRITC (goat anti rabbit, ZF-0317, Beijing ZSGB Biotechnology, 1:200), dried at room temperature and observed via fluorescence microscopy. The sections from mock-treated or HB-01-infected hACE2 mice were directly incubated with FITC-conjugated goat anti-mouse IgG or TRITC-conjugated goat anti-rabbit IgG and used as the negative control. For the expression of human ACE2, the sections from wild-type mice stained with anti-ACE2 antibody were used as the negative control, and the stable cell line expressing human ACE2 was used as the positive control. For the viral antigen, the sections from mock-treated hACE2 mice incubated with anti-S protein antibody were used as the negative control.
Transmission electron microscopy
Supernatant from Vero E6 cell cultures that showed cytopathic effects was collected, inactivated with 2% paraformaldehyde for at least 2 h, and ultracentrifuged to sediment virus particles. The enriched supernatant was negatively stained on film-coated grids for examination. The negative stained grids were observed under transmission electron microscopy.
Statistical analysis
All data were analysed with GraphPad Prism 8.0 software. Statistically significant differences were determined using unpaired t-tests, Student’s t-tests, Welch’s t-tests or Mann–Whitney U-tests, as applicable and according to test requirements. A two-sided P value < 0.05 was considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001. No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
All raw data are available from the corresponding authors on reasonable request. Source data are provided with this paper.
References
Zhu, N. et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020).
Kuba, K. et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11, 875–879 (2005).
Ren, L. L. et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin. Med. J. (Engl.) 133, 1015–1024 (2020).
China National Health Commission. Update on the Novel Coronavirus Pneumonia Outbreak (Jan 24, 2020). http://www.nhc.gov.cn/yjb/s7860/202002/84faf71e096446fdb1ae44939ba5c528.shtml (China National Health Commission, 2020).
Xu, X. et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 63, 457–460 (2020).
Chan, J. F. et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395, 514–523 (2020).
Rivers, T. M. Viruses and Koch’s postulates. J. Bacteriol. 33, 1–12 (1937).
Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020).
de Wit, E., van Doremalen, N., Falzarano, D. & Munster, V. J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 14, 523–534 (2016).
Yang, X. H. et al. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp. Med. 57, 450–459 (2007).
Netland, J., Meyerholz, D. K., Moore, S., Cassell, M. & Perlman, S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J. Virol. 82, 7264–7275 (2008).
Acknowledgements
We thank G. F. Gao for his advice and coordination on this work; H. Deng, X. Yang and L. Zhang for providing the hACE2 mice as a gift; and G. Wong for helping us to proofread the manuscript. This work was supported by National Research and Development Project of China (2020YFC0841100), Fundamental Research Funds for CAMS of China (2020HY320001), National Key Research and Development Project of China (2016YFD0500304), CAMS initiative for Innovative Medicine of China (2016-I2M-2-006) and National Mega projects of China for Major Infectious Diseases (2017ZX10304402, 2018ZX10301403).
Author information
Authors and Affiliations
Contributions
C.Q. and G. Wu conceptualized the study. L.B., W.D., B.H., H.G. and J.L. constructed the methodology. L.B., W.D., B.H., H.G., J.L., L.R., Q.W., P.Y., Y. Xiao, F.Q., Y.Q., F.L., Q.L., W.W., J.X., S.G., M.L., G. Wang, S.W., Z.S., Li Zhao, P.L., Linna Zhao, F.Y., H.W., W. Zhou, N.Z., W. Zhen, H.Y., X.Z., L.G., L.C., C.W., Y.W., X.W., Y. Xu, Q.S., H.L., F.Z., C.M., L.Y., M.Y., J.H., W.X., W.T., X.P. and Q.J. performed the investigations. L.B., J.L., J.X. and Z.S. wrote the original draft, which was reviewed and edited by C.Q. and G. Wu. Funding was acquired by C.Q. and L.B. Resources were provided by C.Q. C.Q. and G. Wu supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Stains of the lungs in HB-01-infected wild-type and hACE2 mice at 3 and 5 dpi.
a, Modified Masson’s trichrome stain of the lung. Compared to the HB-01-infected wild-type mice, increased collagen fibres (blue-stained fibres) in the thickened alveolar interstitium were observed in the HB-01-infected hACE2 mice at 3 and 5 dpi. Blue frames in the top-right corners are magnifications of the region in the corresponding blue box. b, Periodic acid–Schiff (PAS) stain of the respiratory epithelium in bronchioles. A small amount of mucus accumulated on the surface of bronchial epithelial cells. Red frames in the top-right corners are magnifications of the region in the corresponding red box. Scale bars, 40 μm. Data in a, b are representative of three independent experiments.
Extended Data Fig. 2 IHC was carried out to identify MAC2+ macrophages, CD3+ T lymphocytes and CD19+ B lymphocytes.
a, Diffuse infiltration of macrophages (red arrow) in the expanded alveolar septa in HB-01-infected hACE2 mice at 3 and 5 dpi. b, Many T lymphocytes (red arrow) infiltrated the thickened alveolar septa in the first row of b at 5 dpi in the HB-01-infected hACE2 mice. A few B lymphocytes (red arrow) were observed in the HB-01-infected hACE2 mice. Scale bars, 40 μm. Data in a, b are representative of three independent experiments.
Extended Data Fig. 3 Histopathological observations of the organs in HB-01-infected wild-type and hACE2 mice.
There were no substantial histopathological changes in the organs (including myocardium, liver, spleen, kidney, cerebrum, intestine and testis) in HB-01-infected hACE2 mice compared with HB-01-infected wild-type mice. Scale bars, 100 μm. Data are representative of three independent experiments.
Extended Data Fig. 4 IHC observations of the organs in HB-01-infected hACE2 mice.
There were no SARS-CoV-2 antigens in the organs (including myocardium, liver, spleen, kidney, cerebrum, intestine and testis). Scale bars, 50 μm. Data are representative of three independent experiments.
Extended Data Fig. 5 Identification of 7D2 antibody against SARS-CoV-2 S1 protein.
The plate coated with 0.2 μg SARS-CoV-2 S1 protein was incubated with 7D2 antibody as primary antibody (1:200), and detected using HRP-conjugated goat anti-mouse secondary antibody. The titre of antibody was determined using ELISA. Data are mean ± s.d. Significant differences are indicated with asterisks (n = 3, two-tailed unpaired Student’s t-test, **P = 0.0011).
Supplementary information
Supplementary Figure 1
This figure shows the version of Fig. 2 originally published online, which contained a duplication, and the revised, corrected version of Fig. 2.
Rights and permissions
About this article
Cite this article
Bao, L., Deng, W., Huang, B. et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583, 830–833 (2020). https://doi.org/10.1038/s41586-020-2312-y
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41586-020-2312-y
This article is cited by
-
Development and characterization of a fully humanized ACE2 mouse model
BMC Biology (2025)
-
ACE2 shedding exacerbates sepsis-induced gut leak via loss of microbial metabolite 5-methoxytryptophan
Microbiome (2025)
-
Outbreaks of SARS-CoV-2 in minks: public health, environmental and bioethical perspectives
Environmental Sciences Europe (2025)
-
Trends and hotspots in research of virus and gastrointestinal mucosal immunity: a bibliometric analysis of four decades
Virology Journal (2025)
-
Differential patterns of cross-protection against antigenically distinct variants in small animal models of SARS-CoV-2 infection
npj Viruses (2025)




Aariq
I'm VERY concerned about the possible image manipulation in this paper pointed out in this twitter thread: https://twitter.com/Microbi...
Prof Georgy Koentges Replied to Aariq
well spotted. I think the blue boxes are identical but the green and red ones are not.
The reuse of images (across expts) should lead to automatic retraction of the paper. We have no further time to waste on poorly documented and misleading data.
Prof Georgy Koentges
I have put in a critique of this paper yesterday, it was qualified as SPAM first, and is still pending. There are serious technical problems with this paper.
郭平仄 Replied to Prof Georgy Koentges
I saw your comments yesterday. if all the reviewer have question on its technical section? why this can be pulished? if editor ignore the reviwers' concerns, why need peer-review?
Prof Georgy Koentges Replied to 郭平仄
you need to ask them that question. My text has been removed.
I don't care about any type of blame. All we should care about is that information that is published is reliable and appropriately qualified so that others who might not know the necessary details can trust the information to draw public policy conclusion from.
In this case: If the mouse does not show the expected phenotype (expected from humans or similar to that of other experimental systems etc), then people without the necessary technical background might wrongly conclude that animal model systems aren't useful or start to mistrust their own observations. If you do a poorly controlled experiment and find nothing it is not the fault of the experimental system that has tremendous capabilities if used correctly.
Prof Georgy Koentges
I think this paper is deeply flawed and despite the insistence
of the referees the key shortcomings were not fixed in the process.
1.To humanize the mouse model the mouse ACE2 coding sequence needs to be
replaced precisely by the human ACE 2 coding sequence. This has
to be done by gene-targeting to ensure that the new hACE2 expression pattern
and strength exactly corresponds to the normal expression. There is no way
around that and showing an old messy transgene from 2007 is inappropriate. Too
much expression or no expression or a mixture of endogenous mouse ACE2
and transgenic hACE2 expression in space and time can only confound any type of mechanistic analysis. None of this is controlled here. For any transgenic
experiment basic controls need to answer A.where the transgene is expressed, B.
when and by how much, in a permanent transgenic line in direct single cell
comparison with the endogenous gene for each relevant tissue. This determines
the value of the assay system. This wasn’t published by Yang et al 2007 (reference
10) that the authors refer to in their responses. If the transgene is only
picking up part of the overall ACE2 expression pattern or strength in vivo,
then obviously any virus using it would only infect a part of all structures
infected in humans – this explains the current partial phenotype as a trivial
experimental artefact. Nothing can or should be drawn from this poorly controlled experiment about animal or human health. The referees have pointed this out implicitly: why was this not requested as mandatory by the editors?
Prof Georgy Koentges
Part2:All else follows from there:
2. The numbers of experimental animals are inappropriate to draw any conclusion, regardless of the genotype. 3.This paper does not support Koch’s principle for
SARSCov2 and does not add to the mechanistic understanding of viral infection.
Despite the referees’ objections the authors did not amend the text appropriately. 4. H&E as the only analytical method for tissue histopathology in the molecular age belongs into the Palaeozoic, not into a 2020 Nature article: We expect time courses of single cell RNA seq (using mACE2 and transgenic hACE2 as helpful 'baits') throughout the brain and other known affected tissues (brain, kidneys, lung, heart etc) in the corrected mouse transgene. Nothing less.
Peoples’ lives will depend on the reliability and quality of these
experiments that bear the potential to replicate in a few months an entire
lifespan. The mouse as a powerful and appropriate in vivo assay system open to
molecular precision engineering will be at the heart of finding a cure. We
cannot cut corners here.
Kind Regards, Prof Georgy Koentges, School of Life Sciences, University of Warwick
石雨濛 Replied to Prof Georgy Koentges
I have updated this comment. As there are multiple versions of this paper. The reference has changed from "Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China" to "A Novel Coronavirus from Patients with Pneumonia in China, 2019".
Both versions of the paper do not prove Koch's postulate.
The sentence in question:
New Version
The new reference which is '1' in the updated paper:
https://doi.org/10.1056/NEJ...
The following is a quote from the A Novel Coronavirus from Patients with Pneumonia in China, 2019 paper:
* Did not isolate any virus, only obtained genetic material.
* Did not cultivate in host cells, instead used lung cancer cells.
* Did not prove Virus filterability (used only centrifugation).
Old Version
This is the previous version of the paper: https://www.biorxiv.org/con...
I also agree that Koch's postulate has not been satisfied. From this paper.
The references in this sentence is '7' and does not fulfil Koch’s postulate. The reference makes ZERO mention of Koch’s postulate.
https://www.ncbi.nlm.nih.go...
Perhaps they are trying to say Postulate 2 was satisfied but its still unclear.
So reading the “Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China” paper they write:
https://www.ncbi.nlm.nih.go...
Ref[14] http://weekly.chinacdc.cn/e...
Ref 14 is a weekly brief from China CDC. Its not a valid reference suitable for a scientific paper.
As you can see, the Ref 14 turns out to be invalid. Which means any claims regarding isolation in the previous paper are invalid. Which means the original claim to Koch’s postulate is INVALID!!!!
Koch’s postulate requires experimentation. A paper should clearly demonstrate experimentation for each postulate. Its especially dishonest to use a reference for this claim when the reference makes ZERO mention of Koch’s.
Michael Speth - M.S. & B.S. Computer Engineer
ES Replied to 石雨濛
Nowhere in the paragraph you cited from the paper referenced the Lancet paper (Ref. 6). The paragraph cited only Ref. 1 (a NEJM paper) and Ref. 7, an old paper that discusses the applicability of Koch's postulates to viral infection, which is now widely accepted by scientists (and Koch himself) to carry its limitations, particularly Postulate 2 that requires the pathogen to be isolated and grown in pure culture, which taken literally means to grow a pure viral culture. That is biologically impossible given that virus needs a host cell to propagate, unless one accepts that the current tissue culture method of viral cultivation to be sufficient in meeting that requirement, which the authors have shown when quantifying the virus titer using the standard TCID50 assay.
Also, it is not verboten to cite scientific briefs in a scientific paper (in this case, the Ref. 6 Lancet paper that you incorrectly cited) as long as the data in such sources can be vetted. You have not produced any evidence why the brief is invalid/inaccurate only that it's a brief. The brief only announced the release of the complete genome of SARS-CoV-2, the virus isolated from a cluster of pneumonia patients in Wuhan, which is why the sentence in the Lancet paper stating its existence cited the brief as a source, since the brief is the first known source (I believe) documenting the isolation and sequencing of the novel virus.
Based on your logical flow of thinking, the non-invalidation of Ref. 14 in the Lancet paper (again erroneously cited by you and not this article's authors) means that claims to the isolation of the virus and hence, the fulfillment of Koch's postulates still stands. And the paper very well acknowledges that itself and numerous clinical studies collectively fulfill the Koch's postulates, so the authors are not dishonest at all.
I suggest that you temper your claims and your crusade of 'gotcha science debunking' when the scientific consensus and evidence for SARS-CoV-2 being the etiological agent of COVID-19 are very strong.
*Edited on 5/18, 12:40pm to correct the ASM link for Ref. 7
石雨濛 Replied to ES
There are apparently multiple versions of this document. I used the following: https://www.biorxiv.org/con...
Which indeed uses the references I listed for
and
And the reference
So it looks like they changed the reference in the current draft to
So please retract your fallacious claim that I am on a crusade. I clearly debunked a version of this document. Looks like the authors changed the reference in hopes of saving the sentence.
Now for the reference they used. The reference clearly states the following https://www.nejm.org/doi/10...
* Did not isolate any virus, only obtained genetic material.
* Did not cultivate in host cells, instead used lung cancer cells.
* Did not prove Virus filterability (used only centrifugation).
So again, it does not support Koch's postulate. Why are you being so dishonest?
ES Replied to 石雨濛
Again, the authors are consistent in their references for their sentences in both the pre-print Biorxiv paper and the final Nature article here, so you clearly have not debunked any version of the document but only showcased your carelessness in reading scientific papers.
Ref. 7 and Ref. 12 in that pre-printed version are the following:
7. Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497-506, doi:10.1016/s0140-6736(20)30183-5 (2020).
12. Rivers, T. M. Viruses and Koch's Postulates. Journal of bacteriology 33, 1-12 (1937).
which are the corresponding Ref. 1 and Ref. 6 of this paper. So, no, the authors did not change their references to "save" the sentence.
And as for the NEJM paper, you should quote the entire paragraph to give it a proper context given that scientific studies are not viewed in isolation but collectively, which this paper has done in its own pathological observations in an existing (but imperfect) mouse model and clinical observations documented in NEJM. Since then, countless papers have been published fulfilling the Koch's postulates. I suggest that you scour the SARS-Cov-2 literature instead of focusing on "destroying" one paper to disprove all other studies by other experts. I'm putting up the paragraph here so that other readers could be a judge for themselves.
石雨濛 Replied to ES
You are flat out contradicting yourself. You initially wrote:
And when I have explained that the reference changed you claim the following:
I have demonstrated that the reference changed from Lancet's "Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China" to "A Novel Coronavirus from Patients with Pneumonia in China, 2019".
You are clearly being dishonest and contraction yourself.
Just a note, I have updated my original post to include the changes in the references.
ES Replied to 石雨濛
You're right that I did commit the error.
Nonetheless, I stand by my claim that you have not debunked anything but only found a change in source citation, which the authors have the liberty to do so if they think one source is more suited than the other to serve as an example of COVID-19 clinical studies that have been conducted. There's no cardinal rule stating that references in the non-peer-reviewed pre-print must be the same as in the final version: after all, it is a pre-print subject to changes when there are new studies emerging or in this case, when the author has re-reviewed their reference list and found that some references do not belong to certain sentences vs. the other.
Also, one could argue that the authors are being thoughtful and trying to give due credit to the NEJM paper for being the earlier paper (published in January 2020) that linked SARS-CoV-2 to COVID-19 vs. the Lancet paper that was published in February 2020. Another possibility is that the reviewers requested for the change because the NEJM is the first paper (presumably) reporting the potential discovery of the pathogen responsible for the atypical pneumonia outbreak (which it was called back then). Of course, the authors could have cited both papers as examples of clinical studies, why they didn't do that, well it's their prerogative.
Nonetheless, the references cited in both the pre-print and the final version of the paper applies only to the phrase "together with the previous clinical studies": those clinical studies do not stand alone (as the authors in the NEJM paper have clearly stated), and only together with this experimental animal study do they fulfill the Koch's postulates. The authors of this paper have stated that very clearly.
But to accuse them of dishonesty from the get-go instead of giving them the benefit of the doubt they have committed a mistake (or a change instigated by themselves or the reviewers) shows that your intent is less helpful than sowing discord and confusion or distinguishing yourself as a contrarian "debunking" science.
石雨濛 Replied to ES
Those references DO NOT full-fill any of Koch's postulates. I have demonstrated this and you just ignore the facts.
Experimentation is required through the scientific method to prove Koch's postulate. This has NOT been conducted. They are in fact being dishonest because they have made an unsubstantiated claim.
You have no basis for calling my claims 'sowing discord and confusion'.
The scientific method MUST be applied with accurate usage of references. REAL Science demands that claims are substantiated. The scientific method is critical as it requires repeatability. If someone is to prove Koch's they MUST demonstrate how each postulate was met and provide the instructions so that other scientists can repeat the experimentation to confirm that Koch's has been met. It is not sufficient for even 1 paper but many papers to have the same findings.
This is why this paper is flawed. They do not demonstrate through experimentation how Koch's has been met and just make reference to others who DO NOT CLAIM they have met Koch's postulate.
Here are the Koch's postulates as a reminder. A paper must demonstrate that ALL postulates have been met:
1. The microorganism must be found in abundance in all organisms suffering from the disease, but should not be found in healthy organisms.
2. The microorganism must be isolated from a diseased organism and grown in pure culture.
3. The cultured microorganism should cause disease when introduced into a healthy organism.
4. The microorganism must be re-isolated from the inoculated, diseased experimental host and identified as being identical to the original specific causative agent.
ES Replied to 石雨濛
If you have read the peer review file (the link of which is found under the subheading 'Supplementary Information'), you would have seen that referee #3 have raised the question of whether SARS-CoV-2 fulfills the Koch's postulates as modified by Rivers in his 1937 paper, which are widely accepted by the scientific community. (Perhaps it annoys you that the scientific community uses the term 'Koch's postulates' to refer to both his original formulations (which I can't stress enough are limited and have been modified since as we understand microbial pathogenesis much better now than in Koch's years) and the Rivers' modifications for viral pathogens interchangeably, and scientists today almost always refer to the latter. I couldn't care less about your OCD in that regard.) Here are the criteria:
1. Isolation of virus from diseased hosts
2. Cultivation in host cells
3. Proof of filterability
4. Production of comparable disease in the original host species or a related one
5. Re-isolation of the virus
6. Detection of a specific immune response to the virus
Here are referee #3's comments (and mind you, they were from the first review of the paper: it underwent another two rounds of reviews before being published).
And here's the response of the authors to referee #3's comments:
First let's go through criteria 4 to 6, which are experimentally demonstrated here:
Criterion 4. Production of comparable disease in the original host species or a related one
First, the virus strain the authors used in this study is stated below:
Second, Figure 2 shows the gross pathological changes of the infected lung, the histopathological changes (pneumonia and immune cell infiltration), and detection of viral antigens (the spike protein) in the immune cells and airway epithelial cells.
Criterion 5. Re-isolation of the virus
The figure of interest is Fig. 1d. The figure legend for that figure is as follows: "the virus isolated from lungs of ACE2 HB-01 mice at 3 dpi was observed by electron microscope". And reading the Methods section, they used Vero cell, a commonly used monkey kidney epithelial cell line, for the purpose of cultivating viruses to get a sufficient amount of virus particles for electron microscopy examination. Technically less challenging and resource-inexpensive for such a purpose instead of using a primary human cell culture (which requires all sorts of expensive media, growth factors etc.) to maintain such cultures for long periods.
Unless you think that culturing viruses in monkey kidney epithelial cells would significantly change the virus genetically and phenotypically? If so, wouldn't the authors see those changes reflected in their electron microscopy observation? Would their assay for detection of viral nucleic acid still work?
Criterion 6. Detection of a specific immune response to the virus
Figure 1e shows the presence of antibodies (specifically IgG) reactive against the spike protein of SARS-CoV-2 as detected by ELISA.
Now onto criteria 1 to 3, the authors have a paragraph that sums up the fulfillment of those postulates:
This goes back to the NEJM paper and the findings made in the paper that support the authors' claim.
Criterion 1. Isolation of virus from diseased hosts
Criterion 2. Cultivation in host cells
Above is the method that the authors in the NEJM paper used to isolate and culture the virus from the affected patients presenting the respiratory pathology (and appropriate controls served by patients presenting similar pathology but from a site far from the outbreak), thus fulfilling criteria 1 and 2. (Note: they did not use the entire lavage but only the supernatant from the centrifuged samples.)
Contrary to what you claim in your first reply to the original poster, the authors detected more than just genetic material. One, clear cytopathic effects caused by the virus were observed with the infected cell cultures.
Two, you have failed to see in Fig. 3 of the NEJM paper that they directly visualized the viral particles obtained from their cell cultures.
And the method used to obtain those viral particles:
Criterion 2. Cultivation in host cells
Now addressing your concerns with criterion 2 that they did not use 'host cells' but lung cancer cells.
First, using 'host cells' does not mean that one uses cells from the same infected host. When scientists say 'host', they refer to the species in question, and in this case, the host of the virus is human, similar to how we say the host of the Lyme bacterium-carrying tick is the mouse. Because humans are 99.9% genetically similar, there's very little possibility for a virus to demonstrate tropism for only ONE particular individual (unless he/she is a mutant expressing the entry receptor for that particular virus) vs. other human beings. So any human samples would serve the purpose of isolating and cultivating the virus in question.
Many of the cell lines that scientists used are transformed/immortalized human cells, which are pre-cancerous/cancer-like (e.g., HeLa cells). They have been used to elucidate mechanisms in biology because of the inexpensive resources and techniques to maintain such cell cultures. Nowadays with many improvements in cell culture technology and knowledge, primary human cell cultures are gaining more widespread usage but there's one caveat: one has to get them from a living human being and a lot of times, most labs don't have access to clinical facilities that allow procurement of such valuable human samples (and especially during a pandemic). And, primary human cells can't grow indefinitely unless they are immortalized by expressing some oncogenes, for example, as stated earlier. So you could imagine a situation in which one has to consistently perform invasive biopsy (depending on the cells one wants) on human subjects.
In this case, it is conceivable (and accepted) that one uses cells isolated from a human cancer patient as these cancer cells could grow well in conventional, inexpensive, and widely available culture media with less fastidious culture conditions and they could grow long enough for the viral titers to reach a level sufficient for cytopathic effects to manifest.
Of course, we make the assumption here that they obtained cancerous cells from that patient (which is my bet), but it could be that they resected the healthy tissue specimen from that patient's lung for the viral culture.
Criterion 3. Proof of filterability
So now 5 criteria have been fulfilled, what about this one? You correctly stated they performed centrifugation.
This criterion has been stated in various sources but infrequently explicated. The idea is that one has to exclude all conceivable pathogens larger than viruses (using size exclusion membranes) so that only the virus in suspect would remain and examined for pathogenicity. Of course, it'd be better if the NEJM authors elaborate how the centrifugation is performed but here's what possible and why the peer reviewers in that paper (and referee #3 in this paper) didn't raise an alarm about it.
Centrifugation is performed at a certain speed to pellet the larger particles (to simplify matters, major ones include cell debris, bacteria, parasites, fungi), leaving a supernatant that contains mostly viral particles. You could argue that the supernatant would contain other types of viruses that might give rise to the pathology that they are seeing. But as the authors in the NEJM stated in their article (and almost certainly a caveat that'd been picked up by most/all scientists):
They have done a pretty exhaustive exclusion of all known microbial causes and yet-to-be-identified microbes (nucleic acid detection is the most sensitive detection method available thus far since minute quantities of genetic material from live or dead/non-proliferating microbes can be picked up by these tests). Also, if there are other viruses present in the supernatant and they cultured them, the authors would have seen a variety of cytopathic effects in their cell cultures, which is not the case. (Bacterial/fungal/parasites can be directly observed under light microscopy and their growth would cause very different and more gross phenotypic alterations in the cultured cells.) Same goes with the electron microscopy findings in which they would have observed an array of morphologically distinct viruses.
So is criterion 3 fulfilled? Given the consensus of the scientific community (who arguably would easily detect critical gaps in evidence for claims of a new pathogen), I surmise that it has been fulfilled. And, there are other published papers supporting such similar findings (i.e., they detected both the viral nucleic acid and successfully cultured the virus without concurrent detection of other known/unknown microbes). Otherwise, labs around the world would have sent rebuttal letters questioning the validity of those findings. (Unless you presume that thousands of scientists across the globe, sworn in secrecy, are colluding with one another and with the 'mainstream media' to keep this ploy a hush-hush.)
You know, you could have also sent a letter to the NEJM authors asking them to detail their centrifugation protocol?
If these findings do not convince you, may I ask what are your alternative explanations and theories (e.g., exosomes?) for the etiological agent of COVID-19? Could you please cite peer-reviewed experimentally-validated studies to support your claims?
Also don't neglect this study on the experimental infection of macaque with SARS-CoV-2.
Davide Battini Replied to ES
No.
Long story. Good try, but NO you're still there, try again.
Are you joking?
No Koch postulate to date is satisfied, absolutely, and certainly not your points 1, 2, 3.
And I'm talking about both this NEJM study (3 patients) and the second study on NATURE (7 patients) and even the third study on NCBI (1 patient).
Nobody even surpasses them in the simplified version of Rivers' criteria.
After these three studies (or rather, after the first two studies) the whole world does viral "isolates" in the RT-PCR protocol, using standardized primers and probes and kits, fed by the mountains of clone sequences of these studies, yet in fact, not we still really know what kind of pathogen we have before us, the new religion is based on GenBank and the world of Blast.
On CrisprCasXX and the new genomics engineering in VR mode.
Today is not the era of Koch or Rivers.
Today technology can do it easily.
Hundreds of labs purify and filter exosomes every day and as early as the 1980s.
So why don't we do it?
Why don't we ultrafiltrate as god commands for this Mr. X19?
Why don't we see FABULOUS TEM and SEM photos everywhere, pallets or infinite "beds" of filtered viruses that are as clear and indubitable as the sun?
So just to name a few.
Density gradient ultracentrifugation, no.
Host cells and cultures and pure, no.
Reagents and antibiotics as if it rained on precancerous human cultures, and clone, synthetic, animal tissues, yes.
Use (wanted) of HeLa or FRhK4 cells, or Vero E6 animals
just because they are much more reactive to certain "viruses", yes.
RT-PCR test? Better not to talk about it.
There would be much more to sequencing and compulsive sending of genomes
I repeat.
Today is not the era of Koch or Rivers.
Today technology can do it easily.
Ah, finally your criteria 4, 5 and 6 are NOT satisfied.
For example, the same studio that hosts us here on this page is shaking and will have to be withdrawn, because ALL THE PHOTOS AND IMAGES HAVE FAKE PARTS OR ARE WRONG OR HANDLED.
We even have, in Fig. 1d, a giant SARSCoV2 TEM, since it measures about 400nm, excluding spykes.
No way!
See you.
ps: here you can find other manipulated images on this page.
https://twitter.com/Microbi...
https://twitter.com/Microbi...
https://twitter.com/OKallio...