Abstract
Animals such as raccoon dogs, mink and muskrats are farmed for fur and are sometimes used as food or medicinal products1,2, yet they are also potential reservoirs of emerging pathogens3. Here we performed single-sample metatranscriptomic sequencing of internal tissues from 461 individual fur animals that were found dead due to disease. We characterized 125 virus species, including 36 that were novel and 39 at potentially high risk of cross-species transmission, including zoonotic spillover. Notably, we identified seven species of coronaviruses, expanding their known host range, and documented the cross-species transmission of a novel canine respiratory coronavirus to raccoon dogs and of bat HKU5-like coronaviruses to mink, present at a high abundance in lung tissues. Three subtypes of influenza A virus—H1N2, H5N6 and H6N2—were detected in the lungs of guinea pig, mink and muskrat, respectively. Multiple known zoonotic viruses, such as Japanese encephalitis virus and mammalian orthoreovirus4,5, were detected in guinea pigs. Raccoon dogs and mink carried the highest number of potentially high-risk viruses, while viruses from the Coronaviridae, Paramyxoviridae and Sedoreoviridae families commonly infected multiple hosts. These data also reveal potential virus transmission between farmed animals and wild animals, and from humans to farmed animals, indicating that fur farming represents an important transmission hub for viral zoonoses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data reported in this paper have been deposited in the GenBase of the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation (CNCB) under accession numbers C_AA074934.1 to C_AA076255.1. These data are publicly accessible online (https://ngdc.cncb.ac.cn/genbase). The raw sequencing read data have been deposited at the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) of CNCB under the BioProject accession number PRJCA026706. The sequences of four IAVs have been submitted to the GISAID database, and assigned isolate IDs EPI_ISL_19176289, EPI_ISL_19176290, EPI_ISL_19176291 and EPI_ISL_19176294. All multiple-sequence alignments (fasta format), phylogenetic trees and source data related to figures in the main text have been deposited at GitHub (https://github.com/Jin2024-doct/-fur-animals-virus-dataset.git). The map of China is available from the Data Center for Resources and Environmental Sciences at the Chinese Academy of Sciences (http://www.resdc.cn). Source data are provided with this paper.
References
Lindh, E. et al. Highly pathogenic avian influenza A(H5N1) virus infection on multiple fur farms in the south and central Ostrobothnia regions of Finland, July 2023. Eurosurveillance https://doi.org/10.2807/1560-7917.Es.2023.28.31.2300400 (2023).
Enserink, M. Coronavirus rips through Dutch mink farms, triggering culls. Science 368, 1169 (2020).
Peacock, T. P. & Barclay, W. S. Mink farming poses risks for future viral pandemics. Proc. Natl Acad. Sci. USA 120, e2303408120 (2023).
Waller, C. et al. Japanese encephalitis in Australia—a sentinel case. N. Engl. J. Med. 387, 661–662 (2022).
Eledge, M. R., Zita, M. D. & Boehme, K. W. Reovirus: friend and foe. Curr. Clin. Microbiol. Rep. 6, 132–138 (2019).
Sabir, J. S. et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 351, 81–84 (2016).
Woolhouse, M. & Gaunt, E. Ecological origins of novel human pathogens. Crit. Rev. Microbiol. 33, 231–242 (2007).
Taylor, L. H., Latham, S. M. & Woolhouse, M. E. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B 356, 983–989 (2001).
Weingartl, H. M. et al. Genetic and pathobiologic characterization of pandemic H1N1 2009 influenza viruses from a naturally infected swine herd. J. Virol. 84, 2245–2256 (2010).
Lu, M. et al. Zoonotic risk assessment among farmed mammals. Cell 186, 2040 (2023).
He, W. T. et al. Virome characterization of game animals in China reveals a spectrum of emerging pathogens. Cell 185, 1117–1129 (2022).
Vlasova, A. N. et al. Novel canine coronavirus isolated from a hospitalized patient with pneumonia in east Malaysia. Clin. Infect. Dis. 74, 446–454 (2022).
Lednicky, J. A. et al. Independent infections of porcine deltacoronavirus among Haitian children. Nature 600, 133–137 (2021).
Agüero, M. et al. Highly pathogenic avian influenza A(H5N1) virus infection in farmed minks, Spain, October 2022. Eurosurveillance https://doi.org/10.2807/1560-7917.Es.2023.28.3.2300001 (2023).
Conroy, G. Nipah virus outbreak: what scientists know so far. Nature https://doi.org/10.1038/d41586-023-02967-x (2023).
Chua, K. B. et al. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. 4, 145–151 (2002).
Miller, R. S. et al. Cross-species transmission potential between wild pigs, livestock, poultry, wildlife, and humans: implications for disease risk management in North America. Sci. Rep. 7, 7821 (2017).
Magouras, I. et al. Emerging zoonotic diseases: should we rethink the animal–human interface? Front. Vet. Sci. https://doi.org/10.3389/fvets.2020.582743 (2020).
Johnson, C. K. et al. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. Proc. R. Soc. B 287, 20192736 (2020).
Guan, Y. et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302, 276–278 (2003).
Hiono, T. et al. Virological, pathological, and glycovirological investigations of an Ezo red fox and a tanuki naturally infected with H5N1 high pathogenicity avian influenza viruses in Hokkaido, Japan. Virology 578, 35–44 (2023).
Rijks, J. M. et al. Highly pathogenic avian influenza A(H5N1) virus in wild red foxes, the Netherlands, 2021. Emerg. Infect. Dis. 27, 2960–2962 (2021).
Oude Munnink, B. B. et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 371, 172–177 (2021).
Chen, Y. M. et al. Host traits shape virome composition and virus transmission in wild small mammals. Cell 186, 4662–4675 (2023).
Xie, X. T., Kropinski, A. M., Tapscott, B., Weese, J. S. & Turner, P. V. Prevalence of fecal viruses and bacteriophage in Canadian farmed mink (Neovison vison). MicrobiologyOpen 8, e00622 (2019).
Bodewes, R. et al. Viral metagenomic analysis of feces of wild small carnivores. Virol. J. 11, 89 (2014).
Zhang, W. et al. Virome comparisons in wild-diseased and healthy captive giant pandas. Microbiome 5, 90 (2017).
Fenollar, F. et al. Mink, SARS-CoV-2, and the human-animal interface. Front. Microbiol. 12, 663815 (2021).
Ahn, H.-S. et al. Prevalence and genetic features of rabbit hepatitis E virus in Korea. J. Med. Virol. 89, 1995–2002 (2017).
Czudai-Matwich, V., Otte, A., Matrosovich, M., Gabriel, G. & Klenk, H. D. PB2 mutations D701N and S714R promote adaptation of an influenza H5N1 virus to a mammalian host. J. Virol. 88, 8735–8742 (2014).
Wang, W. et al. Divergent Cryptosporidium species and host-adapted Cryptosporidium canis subtypes in farmed minks, raccoon dogs and foxes in Shandong, China. Front. Cell. Infect. Microbiol. 12, 980917 (2022).
Cui, J., Li, F. & Shi, Z. L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 17, 181–192 (2019).
Yang, S. et al. Viral metagenomics reveals diverse viruses in the feces samples of raccoon dogs. Front. Vet. Sci. 8, 693564 (2021).
Lojkić, I. et al. Faecal virome of red foxes from peri-urban areas. Compar. Immunol. Microbiol. Infect. Dis. 45, 10–15 (2016).
Tortorici, M. A. et al. Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein. Cell 185, 2279–2291 (2022).
Lednicky, J. A. et al. Isolation of a novel recombinant canine coronavirus from a visitor to Haiti: further evidence of transmission of coronaviruses of zoonotic origin to humans. Clin. Infect. Dis. 75, e1184–e1187 (2022).
Sah, R., Mohanty, A., Rohilla, R. & Padhi, B. K. A recent outbreak of human H1N2 infection: correspondence. Int. J. Surg. 109, 604–605 (2023).
Szablewski, C. M. et al. Reported global avian influenza detections among humans and animals during 2013-2022: comprehensive review and analysis of available surveillance data. JMIR Publ. Health Surveill. 9, e46383 (2023).
Guillén-Servent, A. & Francis, C. M. A new species of bat of the Hipposideros bicolor group (Chiroptera: Hipposideridae) from Central Laos, with evidence of convergent evolution with Sundaic taxa. Acta Chiropterol. 8, 39–61 (2006).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, D., Liu, C. M., Luo, R., Sadakane, K. & Lam, T. W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
R Core Team. R: a language and environment for statistical computing (R Foundation for Statistical Computing, 2023).
Kolde, R. pheatmap: Pretty Heatmaps. R package version 1.0.12 (R Foundation for Statistical Computing, 2019).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 38, 3022–3027 (2021).
Martin, D. P., Murrell, B., Golden, M., Khoosal, A. & Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 1, vev003 (2015).
Casal, P. E. et al. Evidence for homologous recombination in Chikungunya virus. Mol. Phylogenet. Evol. 85, 68–75 (2015).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Suchard, M. A. et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016 (2018).
Yang, Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39, 306–314 (1994).
Tavaré, S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lectures on Mathematics in the Life Sciences. American Mathematical Society 17, 57–86 (1986).
Hill, V. & Baele, G. Bayesian estimation of past population dynamics in BEAST 1.10 using the Skygrid coalescent model. Mol. Biol. Evol. 36, 2620–2628 (2019).
Acknowledgements
S.S., J.Z., W.W., K.Y., M.L., Z.C., Y.L. and A.H. are financially supported by the National Key Research and Development Program of China (grant no. 2021YFD1801101), the Program of Shanghai Academic Research Leader (23XD1420700), the Young Top-Notch Talents of National Ten Thousand Talent Program; S.S was also supported by the National Natural Science Foundation of China (NSFC grant no. 31922081). W.T.-H. and G.Y. are supported by Young Elite Scientists Sponsorship Program by CAST (2023QNRC001). P.L. and M.A.S. are partially supported by the US National Institutes of Health grant R01 AIAI153044. J.H.-O.P. is supported by the Swedish research council Vetenskapsrådet (grant no. 2020-02593); E.H. by a National Health and Medical Research Council (NHMRC) Investigator award (GNT2017197) and by AIR@InnoHK administered by the Innovation and Technology Commission, Hong Kong Special Administrative Region, China. We acknowledge support from the staff at Advanced Micro Devices, with the donation of parallel computing resources through their AMD HPC Fund used for this research. The Y.B. laboratory was involved in collecting and handling some samples of potential high-risk viruses, such as the HKU5-COV and H5N6 IAV, in accordance with regulations (approved by the Ethics Committee of the Institute of Microbiology, Chinese Academy of Sciences, HP-SQIMCAS2024111), and was financially supported by the National Key Research and Development Program of China (2021YFC2300903). S.S. (alternative email: ssh5658485@163.com) is the lead contact for this paper.
Author information
Authors and Affiliations
Contributions
S.S. and W.T.-H. designed and supervised the research. Y.B., J.Z., X.L. and M.Z. collected samples. J.Z., X.L., M.Z. and G.Y. performed the Sanger sequencing and molecular detection. J.Z., W.W., K.Y., M.L. and W.T.-H. performed the genome assembly, annotation and analysis of abundance. J.Z., P.L., W.T.-H. and S.S. performed the genomic and evolutionary analysis and interpretation. J.H.-O.P., Z.C., Y.L., C.T., A.H., N.H. and M.A.S. assisted in the data interpretation. S.S., W.T.-H., J.Z., J.H.-O.P., E.C.H., Y.B., W.W., K.Y. and M.L. wrote the paper. All of the authors reviewed and edited the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Nathan Grubaugh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
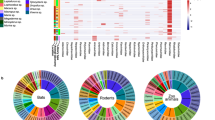
Extended Data Fig. 1 Abundance of vertebrate-associated viruses in fur animals at the species level.
The abundance of each virus was calculated and normalized based on the number of mapped reads per million total reads (RPM) and presented on the Log-10 scale. Different colour blocks represent different types of viruses and source organs. Source data are provided in the Source Data file.
Extended Data Fig. 2 Newly discovered viruses, the infection spectrum of the animals studied and the extent of coinfection.
(a) The infection spectrum of the studied animals, with animals represented by images. Viruses are shown at the nodes, with the node colour specifying the viral family. The size of the nodes represents the number of animals infected by the virus, and the width of the edges indicates the number of libraries of the host infected by the connected virus. (b) The viral families newly identified in the specific host. Each segment of the pie chart corresponds to a distinct animal species, depicted with unique colour, and the donuts with similar lighter colour, signify the newly discovered viral families. (c) Virus co-infection. Viruses are shown at the nodes, with the node colour specifying the viral family. The size of the nodes represents the frequency of co-infections with any other virus, while edge width represents the frequency of co-infections between the two viruses.
Extended Data Fig. 3 Inter-specific phylogenetic trees of 12 major families of vertebrate-associated RNA viruses.
Phylogenetic trees were inferred for each family of RNA viruses based on amino acid sequences of the RNA-dependent RNA polymerase protein. All trees are midpoint-rooted for clarity and display bootstrap values for major branches. Coloured dots represent viruses with different host origins. The scale bar represents the number of amino acid substitutions per site.
Extended Data Fig. 4 Phylogenetic trees of vertebrate-associated RNA viruses from the Flaviviridae and Orthomyxoviridae in fur animals.
Phylogenetic trees of viruses in the (a) Flaviviridae and (b) Orthomyxoviridae were inferred from the amino acid sequences of the RNA-dependent RNA polymerase and hemagglutinin proteins (Orthomyxoviridae). All trees are midpoint-rooted for clarity and display bootstrap values for major branches. Different coloured dots represent viruses with different geographic origins. Colour shading represents different animal orders, and specific species are depicted with animal pictures. The scale bar represents the number of amino acid substitutions per site.
Extended Data Fig. 5 Phylogenetic tree of the Coronaviridae in farmed animals.
Phylogenetic tree of viruses in the Coronaviridae inferred from the amino acid sequences of the RNA-dependent RNA polymerase. The tree is midpoint-rooted for clarity and displays bootstrap values at the major branches. Different coloured dots represent viruses with different geographic origins. Colour shading represents different animal orders, and specific species are depicted with animal pictures. The scale bar represents the number of amino acid substitutions per site.
Extended Data Fig. 6 Inter-specific phylogenetic trees of four vertebrate-associated DNA virus families.
Phylogenetic trees were inferred for each DNA virus family based on the amino acid sequences of conserved viral proteins (DNA viruses = replication related protein, i.e., Anelloviridae: ORF1, Parvoviridae: NS1, Adenoviridae: DNA polymerase, and Circoviridae: Rep protein). All trees are midpoint-rooted for clarity and display bootstrap values for major branches. Coloured dots represent viruses with different host origins. The scale bar represents the number of amino acid substitutions per site.
Extended Data Fig. 7 Intra-specific phylogenetic diversity of multi-host infecting viruses identified in fur animals.
Phylogenetic trees were inferred for each virus species based on the nucleotide sequences of the key gene (i.e., Coronavirus: S1 gene, Paslahepevirus balayani: full genome, Japanese encephalitis virus: E gene, Mammalian orthoreovirus: S1 gene, Norwalk virus: VP1, Rotavirus A: VP7). All trees are midpoint-rooted for clarity and display bootstrap values for the major branches. Coloured dots represent different host sources.
Extended Data Fig. 8 Recombination and phylogenetic analysis of mink-derived Pipistrellus bat coronavirus HKU5.
(a) Maximum clade credibility (MCC) tree based on genome of mink-derived HKU5-like viruses. (b) Simplot was used to perform recombination scanning on the mink-derived HKU5-like sequences and related reference sequences. (c) Neighbour-Net reconstruction based on the complete genome sequences of mink HKU5 and Bat CoVs using Splitstree5, employing the HKY85 substitution model and 1000 bootstraps. (d) IQ-TREE (v2.1.4) was used to estimate maximum likelihood trees based on RdRp and S gene nucleotides, respectively.
Extended Data Fig. 9 Phylogenetic analysis of guinea pig-derived Influenza A virus H1N2.
Maximum clade credibility (MCC) trees based on the HA, MP, NA, NP, NS, PA, PB1, and PB2 gene sequences of H1N2 influenza virus. MCC trees were summarized from Bayesian phylodynamic inferences using BEAST (v1.10.5). Coloured lines and dots represent the host: human (red), rodent (green), and swine (light-blue).
Extended Data Fig. 10 Phylogenetic analysis of two mink-derived Influenza A virus H5N6.
Maximum clade credibility (MCC) trees based on the HA, MP, NA, NP, NS, PA, PB1, and PB2 gene sequences of H5N6. MCC trees were summarized from Bayesian phylodynamic inferences using BEAST (v1.10.5). Different virus clades are depicted in different colours. Blue dots denote the mink-derived H5N6 virus reported here.
Extended Data Fig. 11 Types and abundances of potentially high-risk viruses, along with their geographic and host origins.
(a) The radius of the bubbles indicates the abundance of each potentially high-risk virus, with larger bubbles representing greater abundance. Green bubbles indicate that the virus was identified for the first time in the corresponding host species, while red bubbles indicate previous identification in that host. (b) The relationship between potentially high-risk viruses and their hosts, tissue types, and geographical regions. The line thickness represents the frequency.
Supplementary information
Supplementary Fig. 1
The phylogeny of fur animal hosts, broadening the host range of the viruses identified here, the genome structure of rabbit coronavirus 1 and the phylogenetic tree of H6N2 influenza A virus. a, The phylogeny and taxonomic relationships of fur animal hosts surveyed and related representative mammalian species (data from TimeTree; https://timetree.org/). The dots indicate species that are involved in this study, with a star highlighting the phylogenetic position of humans. Owing to a lack of data for the species Notamacropus rufogriseus, the taxonomic relationships from the closely related Notamacropus parma were used. b, The heat map shows the expanded animal host range for 93 viruses, and the stacked bar chart on the right shows the number of expanded host for each virus. Different hosts are distinguished by colour. c, The genomic structure of the six rabbit CoV-1 sequences and the reference sequence of Pika coronavirus. d, The maximum likelihood tree and lineage classification of the HA gene of the H6N2 virus identified in muskrat.
Supplementary Table 1
Virus species demarcation criteria used in this study, related to the Methods.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, J., Wan, W., Yu, K. et al. Farmed fur animals harbour viruses with zoonotic spillover potential. Nature 634, 228–233 (2024). https://doi.org/10.1038/s41586-024-07901-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-024-07901-3
This article is cited by
-
Ancient DNA insights into diverse pathogens and their hosts
Nature Reviews Genetics (2026)
-
Clinical metagenomics for diagnosis and surveillance of viral pathogens
Nature Reviews Microbiology (2026)
-
Viral emergence and pandemic preparedness in a One Health framework
Nature Reviews Microbiology (2026)
-
Canine distemper virus surveillance and sequencing in invasive feral American mink (Neogale vison) in Poland
Biological Invasions (2026)
-
The silent spread: uncovering the diversity and evolution of poxviruses in ticks across Western China’s host landscapes
Virology Journal (2025)