Abstract
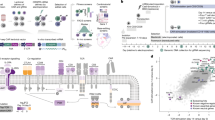
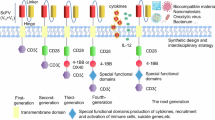
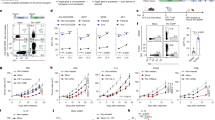
Chimeric antigen receptor (CAR) T cells are highly effective in haematological malignancies1. However, progressive loss of CAR T cells contributes to relapse in many patients2,3,4. Here we performed in vivo loss-of-function CRISPR screens in CAR T cells targeting B cell maturation antigen to investigate genes that influence CAR T cell persistence and function in a human multiple myeloma model. We tracked the expansion and persistence of CRISPR library-edited T cells in vitro and at early and late time points in vivo to track the performance of gene-modified CAR T cells from manufacturing to survival in tumours. The screens revealed context-specific regulators of CAR T cell expansion and persistence. Ablation of RASA2 and SOCS1 enhanced T cell expansion in vitro, whereas loss of PTPN2, ZC3H12A and RC3H1 conferred early growth advantages to CAR T cells in vivo. Notably, we identified cyclin-dependent kinase inhibitor 1B (encoded by CDKN1B), a cell cycle regulator, as the most important factor limiting CAR T cell fitness at late time points in vivo. CDKN1B ablation increased CAR T cell proliferation and effector function, significantly enhancing tumour clearance and overall survival. Our findings reveal differing effects of gene perturbation on CAR T cells over time and in different environments, highlight CDKN1B as a promising target to generate highly effective CAR T cells for multiple myeloma and underscore the potential of in vivo screening for identifying genes to enhance CAR T cell efficacy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Single-cell and bulk transcriptomic data are available through GEO at GSE302503 and GSE302504. Source data are provided with this paper.
References
June, C. H., O’Connor, R. S., Kawalekar, O. U., Ghassemi, S. & Milone, M. C. CAR-T cell immunotherapy for human cancer. Science 359, 1361–1365 (2018).
Rodriguez-Otero, P. et al. Ide-cel or standard regimens in relapsed and refractory multiple myeloma. N. Engl. J. Med. 388, 1002–1014 (2023).
San-Miguel, J. et al. Cilta-cel or standard care in lenalidomide-refractory multiple myeloma. N. Engl. J. Med. 389, 335–347 (2023).
Ruella, M., Korell, F., Porazzi, P. & Maus, M. V. Mechanisms of resistance to chimeric antigen receptor-T cells in haematological malignancies. Nat. Rev. Drug Discov. 22, 976–995 (2023).
Munshi, N. C. et al. Idecabtagene vicleucel in relapsed and refractory multiple myeloma. N. Engl. J. Med. 384, 705–716 (2021).
Berdeja, J. G. et al. Ciltacabtagene autoleucel, a B-cell maturation antigen-directed chimeric antigen receptor T-cell therapy in patients with relapsed or refractory multiple myeloma (CARTITUDE-1): a phase 1b/2 open-label study. Lancet 398, 314–324 (2021).
Schmidt, R. et al. CRISPR activation and interference screens decode stimulation responses in primary human T cells. Science 375, eabj4008 (2022).
Wu, J. E. et al. In vitro modeling of CD8+ T cell exhaustion enables CRISPR screening to reveal a role for BHLHE40. Sci. Immunol. 8, eade3369 (2023).
Shang, W. et al. Genome-wide CRISPR screen identifies FAM49B as a key regulator of actin dynamics and T cell activation. Proc. Natl Acad. Sci. USA 115, E4051–E4060 (2018).
Simeonov, D. R. & Marson, A. CRISPR-based tools in immunity. Annu. Rev. Immunol. 37, 571–597 (2019).
Wang, D. et al. CRISPR screening of CAR-T cells and cancer stem cells reveals critical dependencies for cell-based therapies. Cancer Discov. 11, 1192–1211 (2021).
Shifrut, E. et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell 175, 1958–1971 (2018).
Carnevale, J. et al. RASA2 ablation in T cells boosts antigen sensitivity and long-term function. Nature 609, 174–182 (2022).
Freitas, K. A. et al. Enhanced T cell effector activity by targeting the Mediator kinase module. Science 378, eabn5647 (2022).
Chen, Z. et al. In vivo CD8+ T cell CRISPR screening reveals control by Fli1 in infection and cancer. Cell 184, 1262–1280 (2021).
Wei, J. et al. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature 576, 471–476 (2019).
Lynn, R. C. et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 576, 293–300 (2019).
Friedman, K. M. et al. Effective targeting of multiple B-cell maturation antigen-expressing hematological malignances by anti-B-cell maturation antigen chimeric antigen receptor T cells. Hum. Gene Ther. 29, 585–601 (2018).
Stock, S., Schmitt, M. & Sellner, L. Optimizing manufacturing protocols of chimeric antigen receptor T cells for improved anticancer immunotherapy. Int. J. Mol. Sci. 20, 6223 (2019).
Locke, F. L. et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1–2 trial. Lancet Oncol. 20, 31–42 (2019).
Xu, Y. et al. Closely related T-memory stem cells correlate with in vivo expansion of CAR.CD19-T cells and are preserved by IL-7 and IL-15. Blood 123, 3750–3759 (2014).
Cieri, N. et al. IL-7 and IL-15 instruct the generation of human memory stem T cells from naive precursors. Blood 121, 573–584 (2013).
Dwyer, C. J. et al. Fueling cancer immunotherapy with common γ chain cytokines. Front. Immunol. 10, 263 (2019).
Abou-El-Enein, M. et al. Scalable manufacturing of CAR T cells for cancer immunotherapy. Blood Cancer Discov. 2, 408–422 (2021).
Amatya, C. et al. Optimization of anti-CD19 CAR T cell production for treatment of patients with chronic lymphocytic leukemia. Mol. Ther. Methods Clin. Dev. 32, 101212 (2024).
Lane-Reticker, S. K. et al. Protocol for in vivo CRISPR screening using selective CRISPR antigen removal lentiviral vectors. STAR Protoc. 4, 102082 (2023).
Liau, N. P. D. et al. The molecular basis of JAK/STAT inhibition by SOCS1. Nat. Commun. 9, 1558 (2018).
Sporri, B., Kovanen, P. E., Sasaki, A., Yoshimura, A. & Leonard, W. J. JAB/SOCS1/SSI-1 is an interleukin-2-induced inhibitor of IL-2 signaling. Blood 97, 221–226 (2001).
Flosbach, M. et al. PTPN2 deficiency enhances programmed T cell expansion and survival capacity of activated T cells. Cell Rep. 32, 107957 (2020).
Simoncic, P. D., Lee-Loy, A., Barber, D. L., Tremblay, M. L. & McGlade, C. J. The T cell protein tyrosine phosphatase is a negative regulator of Janus family kinases 1 and 3. Curr. Biol. 12, 446–453 (2002).
Matsushita, K. et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458, 1185–1190 (2009).
Mino, T. et al. Regnase-1 and roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell 161, 1058–1073 (2015).
Behrens, G. et al. Disrupting roquin-1 interaction with regnase-1 induces autoimmunity and enhances antitumor responses. Nat. Immunol. 22, 1563–1576 (2021).
Uehata, T. et al. Malt1-induced cleavage of regnase-1 in CD4+ helper T cells regulates immune activation. Cell 153, 1036–1049 (2013).
Kloss, C. C. et al. Dominant-negative TGF-β receptor enhances PSMA-targeted human CAR T cell proliferation and augments prostate cancer eradication. Mol. Ther. 26, 1855–1866 (2018).
Larson, R. C. et al. Anti-TACI single and dual-targeting CAR T cells overcome BCMA antigen loss in multiple myeloma. Nat. Commun. 14, 7509 (2023).
Lemoine, J., Ruella, M. & Houot, R. Overcoming intrinsic resistance of cancer cells to CAR T-cell killing. Clin. Cancer Res. 27, 6298–6306 (2021).
Razavipour, S. F., Harikumar, K. B. & Slingerland, J. M. p27 as a transcriptional regulator: new roles in development and cancer. Cancer Res. 80, 3451–3458 (2020).
Harrison, S. J. et al. CAR+ T-cell lymphoma post ciltacabtagene autoleucel therapy for relapsed refractory multiple myeloma. Blood 142, 6939 (2023).
Perica, K. et al. CD4+ T-cell lymphoma harboring a chimeric antigen receptor integration in TP53. N. Engl. J. Med. 392, 577–583 (2025).
Ghilardi, G. et al. T cell lymphoma and secondary primary malignancy risk after commercial CAR T cell therapy. Nat. Med. 30, 984–989 (2024).
Larson, R. C. & Maus, M. V. Recent advances and discoveries in the mechanisms and functions of CAR T cells. Nat. Rev. Cancer 21, 145–161 (2021).
Zheng, W. et al. Regnase-1 suppresses TCF-1+ precursor exhausted T-cell formation to limit CAR-T-cell responses against ALL. Blood 138, 122–135 (2021).
LaFleur, M. W. et al. A CRISPR–Cas9 delivery system for in vivo screening of genes in the immune system. Nat. Commun. 10, 1668 (2019).
LaFleur, M. W. et al. PTPN2 regulates the generation of exhausted CD8+ T cell subpopulations and restrains tumor immunity. Nat. Immunol. 20, 1335–1347 (2019).
Lin, C.-P. et al. Multimodal stimulation screens reveal unique and shared genes limiting T cell fitness. Cancer Cell 42, 623–645 (2024).
Li, R. et al. Comparative optimization of combinatorial CRISPR screens. Nat. Commun. 13, 2469 (2022).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Acknowledgements
F.K. received funding from the German Research Foundation (DFG; 466535590). G.E. received funding from the Gilead Sciences Research Scholar Program. D.S.-B. received funding from the CRIS Foundation Out-Back Fellowship Programme (outback2021_6) and from the Spanish Society of Medical Oncology. F.B. received funding from the American–Italian Cancer Foundation and the Italian Association for Cancer Research. This work was funded by NIH R01 CA238268 (M.V.M.) and the Krantz Breakthrough Award.
Author information
Authors and Affiliations
Contributions
CRISPR screen conception and experimental design: N.H.K., F.K., T.K., R.C.L., J.G.D., D.S., K.B.Y., R.T.M. and M.V.M. Performed CRISPR screen experiments: N.H.K., F.K., T.K., A.Y.C., A.B., S.G., H.W.P., M. Pezeshki, A.R. and J.S.M.T.S. Target validation and mechanistic experiments: N.H.K., G.E., F.K., T.K., A.Y.C., M.Z., A.B., A.A., M.C.K., S.G., H.W.P., M. Pezeshki, A.R., J.S.M.T.S., M. Phillips, S.P., D.S.-B., E.P.D. and F.B. Animal handling was led by A.B. Analysis and interpretation of data: N.H.K., G.E., F.K., T.K., C.N., S.A., A.Y.C., M.Z., A.B., A.A., M.C.K., S.G., H.W.P., M. Pezeshki, A.R., J.S.M.T.S., M. Phillips, F.B., M.B.L., R.C.L., J.G.D., D.S., K.B.Y., R.T.M. and M.V.M. Computational analyses: C.N., S.A. and A.Y.C. Manuscript writing and revision: N.H.K., G.E., F.K., C.N., T.R.B., K.B.Y., R.T.M. and M.V.M.
Corresponding authors
Ethics declarations
Competing interests
M.V.M., N.H.K., F.K., T.R.B. and R.T.M. are inventors on patents filed by MGH and the Broad Institute on these technologies. M.V.M. is an inventor on patents related to adoptive cell therapies, held by MGH (some licensed to ProMab and Luminary) and the University of Pennsylvania (some licensed to Novartis). M.V.M. holds equity in 2seventy bio, A2 Bio, AffyImmune, BendBio, Cargo, GBM newco, Model T bio, NexImmune and Oncternal. M.V.M. receives grant and/or research support from Kite Pharma, Moderna and Sobi. M.V.M. has served as a consultant for multiple companies involved in cell therapies. M.V.M.’s competing interests are managed by Mass General Brigham. R.T.M. has received consulting or speaking fees from Bristol Myers Squibb, Gilead Sciences and Immunai Therapeutics, has equity ownership in OncoRev and receives research funding from Calico Life Sciences. M.B.L. is an inventor on patents related to adoptive cell therapies held by MGH and has served as a consultant for BioNTech and Cabaletta Bio. M.B.L. holds equity in AbbVie. R.C.L. is an inventor on patents related to adoptive cell therapies held by MGH, has served as a consultant for Cargo and is now an employee of Link Cell Therapies. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Joseph Fraietta and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 In vivo human CAR-T cell screening results are comparable across multiple healthy human donors.
a. Construct designs for the 4-1BB BCMA CAR (pCAR) and double-guide cassette (pGuide) containing the Mario sgRNA library (variable sgRNA). b. Timeline of the MM.1S stress model with 21 day tumor engraftment and 2E6 CAR treatment. Tumor growth was tracked by BLI. Data presented as mean +/– SEM. n = 3 mice (tumor only, Mario library (no CAR)) and n = 5 (CAR-T, Mario-CAR-T) per group from one healthy donor (ND202). Statistical significance amongst groups at day 35 as measured by two-way ANOVA with Tukey’s multiple comparison. c. CD3 expression after day -5 CD3 negative enrichment (ND216, ND99, ND106). d. Representative Mario-CAR-T cell staining for CD34 indicating CAR transduced cells and NGFR for sgRNA library transduced cells from ND216. e. Replicate autocorrelation analysis scatter plots. Pearson’s correlations are calculated for the library distribution of one animal versus any other animal, two averaged animals versus any other two, and so on. The mean of all possible combinations is plotted. f. Quantification of replicate autocorrelation analysis. Pearson’s correlations are calculated for the library distribution of one animal versus any other animal, two averaged animals versus any other two, and so on. The mean of all possible combinations is plotted. g. z-scored abundance histograms of gene targeting or intergenic control sgRNAs across screening time points and conditions. h. Donor Pearson correlations across screening time points and conditions. ns: non significant.
Extended Data Fig. 2 In vivo screens identify genes that modify CAR-T cell abundance and transcriptional phenotype.
a. Genes ranked by Log2(fold change) during in vitro manufacturing in IL-7/IL-15 (end-of-production vs. baseline, left panel) and early in vivo (day 7 vs. end-of-production, middle panel) or late in vivo (day 21 vs. end-of-production, right panel). Enriched genes are shown in red and depleted genes are shown in blue with circle size corresponding to –log10 (FDR). n = 2 healthy donor T cells (ND106, ND216). b. Frequency histograms of enrichment or depletion of sgRNAs for known T cell regulatory genes, grouped by respective period. In vitro manufacturing was performed in IL-2. n = 3 healthy donor T cells (ND216, ND99, ND106) c. CD4 and CD8 T cell composition (left panel) and phenotype (middle and right panels) at the beginning of CAR-T cell production (day 0) or at the EOP (day 13) from three healthy donors. Data are presented as mean +/– SEM. Naïve (CD45RA + CCR7 + CD95-), T memory stem cells (TSCM: CD45RA + CCR7 + CD95+), central memory (CM: CD45RA-CCR7+), effector memory (EM: CD45RA-CCR7-), terminally differentiated effector memory (TEMRA: CD45RA + CCR7-) T cells. One-way ANOVA, with Sidak’s multiple comparison test (statistical analysis is calculated between IL-2 and IL-7/IL-15 conditions for each genetic perturbation). CM: intergenic IL2 vs IL7/15 **p = 0.0063, PTPN2 KO IL2 vs IL7/15 **p = 0.0067, CDKN1B KO IL2 vs IL7/15 **p = 0.0072; TEMRA: intergenic IL2 vs IL7/15 ***p = 0.0002, PTPN2 KO IL2 vs IL7/15 ***p = 0.0001, CDKN1B KO IL2 vs IL7/15 ***p = 0.0001. d. Abundance of sgRNAs targeting individual genes across the entire screen workflow for Mario-CAR-T cells produced in IL-7/IL-15.
Extended Data Fig. 3 Analysis and validation of knockout CAR-T cells.
a. Proportions of each knockout cell type amongst clusters. b. Hallmark gene set enrichment analysis (GSEA) of pseudo-bulk pooled PTPN2 KO (top panel) or CDKN1B KO (bottom panel) CAR-T cells compared to no guide CAR-T cells. c. Construct design for the 4-1BB BCMA CAR (pCAR) and double-guide cassette (pGuide) containing individual gene sgRNAs (variable sgRNA): CDKN1B, IL2RA, PTPN2, and RASA2. d. Quantification of insertions/deletions (indels) in sgRNA target region by next generation sequencing. Data represents target sgRNA 1 and 2 for CDKN1B, PTPN2, and RASA2, respectively (n = 2 samples); presented as mean +/– SEM. e. IL2RA expression by flow cytometry, with IL2RA KO CAR-T cells (before electroporation/CD3 negative selection and after) compared to UTD and intergenic control KO (both IL-2 and IL-7/IL-15-produced, respectively). UTD = untransduced T cells.
Extended Data Fig. 4 In vitro cytotoxicity of knockout CAR-T cells.
a-b. 16-hour luciferase-based killing assay of knockout CAR-T cells co-cultured with MM.1S tumor cells (a, n = 4 technical replicates from CAR-T cells generated from two normal donors, ND116, ND202) or RPMI-8226 (b, n = 2 technical replicates from CAR-T cells generated from one normal donor), at different effector to target (E:T) ratios (from 10:1 to 1:100). Upper panels display comparison of intergenic KO CAR-T cells to UTD. Middle and lower panels show relative killing of IL2RA KO (IL-2- or IL-7/IL-15-produced; middle panel) or CDKN1B KO, PTPN2 KO, and RASA2 KO (lower panel) compared to intergenic KO CAR-T cells. Data presented as mean +/– SEM. Statistical significance was measured by two-way ANOVA with Tukey’s multiple comparison test. UTD: untransduced T cells. ns: non significant.
Extended Data Fig. 5 CDKN1B ablation increases the efficacy of BCMA CAR-T cells in vivo.
a. MM.1S tumor burden as measured by bioluminescent imaging (BLI) of mice treated with intergenic control KO or CDKN1B KO BCMA CAR-T cells. NSG mice were injected intravenously with 1E6 MM.1S followed by transfer of CAR-T cells 21 days later. n = 3 mice per group from one healthy donor (ND202). Data presented as mean +/– SEM. Statistical significance was measured compared to the intergenic KO CAR-T cell treated group at day 77 (CDKN1B KO) as measured by two-way ANOVA with Tukey’s multiple comparison test. b. Overall survival of mice treated with intergenic control KO or CDKN1B KO BCMA CAR-T cells. n = 13 for tumor only and intergenic control KO CAR-T cell treated animals and n = 16 for CDKN1B KO CAR-T cell treated animals. Data were combined from multiple experiments using T cells from two healthy human donors (ND202, ND116) and two sgRNA per gene target. Statistical significance was measured by log-rank (Mantel–Cox test) for Kaplan–Meier curves. c. Representative flow plot and quantification of the number of BCMA molecules expressed on the indicated tumor cell lines.
Extended Data Fig. 6 CDKN1B ablation improves the expansion and persistence of BCMA CAR-T cells from multiple myeloma patient-1 in the MM.1S tumor model.
a. MM.1S tumor burden as measured by bioluminescent imaging (BLI) of mice treated with intergenic control KO or CDKN1B KO BCMA CAR-T cells. NSG mice were injected intravenously with 1E6 MM.1S followed by transfer of CAR-T cells (from MM patient 1) 21 days later. n = 8 mice per group for intergenic control KO or CDKN1B KO groups. n = 5 mice in the tumor only group. On day 66 post-CAR-T cell treatment, mice were re-challenged with 4E6 MM.1S cells (1st re-challenge), and again on day 84 with 8E6 MM.1S cells (2nd re-challenge). At each re-challenge, naïve mice (n = 3) were injected as controls. Data presented as mean +/– SEM. b-c. Percentage (b) and numbers (c) (mean +/– SEM, n = 8 mice per group) of intergenic control KO or CDKN1B KO CAR-T cells in the peripheral blood at the indicated time post CAR-T cell infusion. Two-way ANOVA with Tukey’s multiple comparison test. d. CD8/CD4 CAR-T cells ratio (mean +/– SEM, n = 8 mice per group) in the peripheral blood of the mice at the indicated time post CAR-T cell infusion. Two-way ANOVA test. e-f. Phenotype of intergenic control KO or CDKN1B KO CD4+ and CD8+ CAR-T cells at EOP (e) or in the mice (f, mean +/– SEM, n = 8 mice per group) at the indicated time post CAR-T cell infusion. T memory stem cells, (TSCM, CD45RA + CCR7 + CD95+), central memory (CM, CD45RA-CCR7+), effector memory (EM, CD45RA-CCR7-) and terminally differentiated effector memory (TEMRA, CD45RA + CCR7-) T cells. Statistical significance measured by two-sided Student’s t-test. Day14: CD4 + CAR-T: TSCM **p = 0.0015, EM **p = 0.001, TEMRA *p = 0.0183; CD8 + CAR-T: TSCM ****p < 0.0001, EM ****p < 0.0001. Day 21: CD4 + CAR-T: CM *p = 0.03, TEMRA p = 0.0022; CD8 + CAR-T: TSCM ****p < 0.0001, CM *p = 0.016, EM **p = 0.0072, TEMRA *p = 0.0007; Day 28: CD4 + CAR-T: CM **p = 0.0025, TEMRA p = 0.0007; CD8 + CAR-T: TSCM **p = 0.0012, EM **p = 0.002, TEMRA ****p < 0.0001; Day35: CD4 + CAR-T: TSCM *p = 0.0157, CM **p = 0.0069, TEMRA ****p < 0.0001; CD8 + CAR-T: TSCM ***p = 0.0006, EM **p = 0.001, TEMRA **p = 0.0076. g. Percentage (mean +/– SEM, n = 8 mice per group) of PD1, TIM3 and CD39 expression in intergenic control KO or CDKN1B KO CD4+ and CD8+ CAR-T cells in the peripheral blood of the mice at the indicated time post CAR-T cell infusion. Two-way ANOVA with Tukey’s multiple comparison test. h) MM.1S tumor burden (mean +/– SEM, n = 8 mice per group) as measured by BLI on day 15 post 1st re-challenge. Each dot represents an individual mouse. i) Number of CAR-T cells (mean +/– SEM, n = 8 mice per group) in the blood of mice treated with intergenic control or CDKN1B KO CAR-T cells, measured 10 days before and 14 days after the 1st tumor re-challenge. Statistical significance measured by two-sided Student’s t-test. j) MM.1S tumor burden (mean +/– SEM, n = 8 mice per group) as measured by BLI on day 41 post 2nd tumor re-challenge. Each dot represents an individual mouse.
Extended Data Fig. 7 CDKN1B ablation improves the expansion and persistence of BCMA CAR-T cells from multiple myeloma patient-2 in the MM.1S tumor model.
a. MM.1S tumor burden as measured by bioluminescent imaging (BLI) of mice treated with intergenic control KO or CDKN1B KO BCMA CAR-T cells. NSG mice were injected intravenously with 1E6 MM.1S followed by transfer of CAR-T cells (from MM patient 2) 21 days later. n = 6 mice per group. On day 71 post-CAR-T cell treatment, mice were re-challenged with 8E6 MM.1S cells. Naïve mice (n = 3) were injected as controls. Data presented as mean +/– SEM. b-c. Percentage (b) and numbers (c) (mean +/– SEM, n = 6 mice per group) of intergenic control KO or CDKN1B KO CAR-T cells the peripheral blood at the indicated time post CAR-T cell infusion. Two-way ANOVA with Tukey’s multiple comparison test. d. CD8/CD4 CAR-T cells ratio (mean +/– SEM, n = 6 mice per group) over time in the peripheral blood of the mice. e-f. Phenotype of intergenic control KO or CDKN1B KO CD4+ and CD8+ CAR-T cells at EOP (e) or in the mice (f, mean +/– SEM, n = 6 mice per group) at the indicated time post CAR-T cell infusion. T memory stem cells, (TSCM, CD45RA + CCR7 + CD95+), central memory (CM, CD45RA-CCR7+), effector memory (EM, CD45RA-CCR7-) and terminally differentiated effector memory (TEMRA, CD45RA + CCR7-) T cells. Statistical significance measured by two-sided Student’s t test. Day 14: CD4 + CAR-T: TEMRA *p = 0.012 g. Percentage (mean +/– SEM, n = 6 mice per group) of PD1, TIM3 and CD39 expression in intergenic control KO or CDKN1B KO CD4+ and CD8+ CAR-T cells in the peripheral blood of the mice at the indicated time post CAR-T cell infusion. Two-way ANOVA with Tukey’s multiple comparison test. h) MM.1S tumor burden (mean +/– SEM, n = 6 mice per group) as measured by BLI on day 41 post re-challenge. Each dot represents an individual mouse.
Extended Data Fig. 8 CDKN1B ablation improves the expansion and persistence of BCMA CAR-T cells from multiple myeloma patient-1 in the RPMI-8226 tumor model.
a. RPMI-8226 tumor burden as assessed by caliper measurements of mice treated with intergenic control KO or CDKN1B KO BCMA CAR-T cells. NSG mice were implanted subcutaneously with 5E6 RPMI-8226 followed by transfer of CAR-T cells (from MM patient 1) 14 days later. n = 7 mice in tumor only and control sgRNA groups; n = 6 mice in CDKN1B KO group. Data presented as mean +/– SEM. b-c. Percentage (b) and numbers (c) (mean +/– SEM) of intergenic control KO (n = 7 mice) or CDKN1B KO (n = 6 mice) CAR-T cells the peripheral blood at the indicated time post CAR-T cell infusion. Two-way ANOVA with Tukey’s multiple comparison test. d. CD8/CD4 CAR-T cells ratio (mean +/– SEM) in the peripheral blood of intergenic control KO (n = 7 mice) or CDKN1B KO (n = 6 mice) mice at the indicated time post CAR-T cell infusion. e-f. Phenotype of intergenic control KO (n = 7 mice) or CDKN1B KO (n = 6 mice) CD4+ and CD8+ CAR-T cells at EOP (e) or in the mice (f) at the indicated time post CAR-T cell infusion. T memory stem cells, (TSCM, CD45RA + CCR7 + CD95+), central memory (CM, CD45RA-CCR7+), effector memory (EM, CD45RA-CCR7-) and terminally differentiated effector memory (TEMRA, CD45RA + CCR7-) T cells. Statistical significance measured by two-sided Student’s t-test. Day7: CD4 + CAR-T: CM *p = 0.0464, EM *p = 0.021; Day14: CD4 + CAR-T: CM *p = 0.041, EM *p = 0.046; CD8 + CAR-T: CM **p = 0.0033; Day21: CD4 + CAR-T: CM ***p = 0.0004, TEMRA *p = 0.0141; CD8 + CAR-T: CM ****p < 0.0001, EM *p = 0.0138, TEMRA *p = 0.0267; Day28: CD4 + CAR-T: CM **p = 0.0041, TEMRA **p = 0.0038; CD8 + CAR-T: CM ****p < 0.0001, TEMRA ***p = 0.0002. g. Percentage (mean +/– SEM) of PD1, TIM3 and CD39 expression in intergenic control KO (n = 7 mice) or CDKN1B KO (n = 6 mice) CD4+ and CD8+ CAR-T cells in the peripheral blood of the mice at the indicated time post CAR-T cell infusion. Two-way ANOVA with Tukey’s multiple comparison test.
Extended Data Fig. 9 CDKN1B ablation increases the proliferation of BCMA CAR-T cells.
a. Comparison of cell cycle (G0/G1, S, G2/M) between CDKN1B KO and intergenic control KO CAR-T cells. Stimulation with irradiated K562-BCMA tumor cells was conducted on day 0 and day 6, with flow cytometry measurements on day 0 (before stimulation), day 1, day 7, and day 14 (schematic overview, left). *p < 0.05, and **p < 0.01.
Extended Data Fig. 10 CDKN1B ablation improves the expansion of T cells engineered with a cilta-cel-like BCMA-targeting CAR.
a. MM.1S tumor burden as assessed by bioluminescence imaging (BLI) of mice treated with intergenic control KO or CDKN1B KO BCMA cilta-cel-like CAR-T cells. NSG mice were injected intravenously with 1E6 MM.1S followed by transfer of CAR-T cells (ND116) 21 days later. n = 5 mice in tumor only and control KO groups; n = 6 mice in CDKN1B KO group. b. Survival curve, n = 5 mice in tumor only and control sgRNA groups; n = 6 mice in CDKN1B sgRNA 1 group per group. c-d. Percentage (c) and numbers (d) (mean +/– SEM) of intergenic control KO (n = 5 mice) or CDKN1B KO (n = 6 mice) CAR-T cells in the peripheral blood at the indicated time post CAR-T cell infusion. Two-way ANOVA with Tukey’s multiple comparison test. e. MM.1S tumor burden (mean +/– SEM, n = 6 mice in tumor only and CDKN1B sgRNA 1 groups; n = 5 mice in intergenic control KO group) as assessed by BLI in mice treated with intergenic control KO or CDKN1B KO BCMA cilta-cel-like CAR-T cells (ND202). f-g. Percentage (f) and numbers (g) (mean +/– SEM, n = 6 mice in CDKN1B sgRNA 1 group; n = 5 mice in control sgRNA group) of intergenic control KO or CDKN1B KO CAR-T cells the peripheral blood at the indicated time post CAR-T cell infusion. Two-way ANOVA with Tukey’s multiple comparison test. h. CD8/CD4 CAR-T cells ratio (mean +/– SEM, n = 6 mice in CDKN1B KO group; n = 5 mice in intergenic control KO group) over time in the peripheral blood of the mice. i-j. Phenotype of intergenic control KO or CDKN1B KO (n = 6 mice) CD4+ and CD8+ CAR-T cells at EOP (i) or in the mice (j, mean +/– SEM, n = 6 mice in CDKN1B sgRNA 1 group; n = 5 mice in control sgRNA group) at the indicated time post CAR-T cell infusion. T memory stem cells, (TSCM, CD45RA + CCR7 + CD95+), central memory (CM, CD45RA-CCR7+), effector memory (EM, CD45RA-CCR7-) and terminally differentiated effector memory (TEMRA, CD45RA + CCR7-) T cells. Statistical significance measured by two-sided Student’s t-test. Day21: CD8 + CAR-T: TSCM *p = 0.0138; Day28: CD4 + CAR-T: TEMRA *p = 0.0328. k. Percentage (mean +/– SEM) of PD1, TIM3 and CD39 expression in intergenic control KO (n = 5 mice) or CDKN1B KO (n = 6 mice) CD4+ and CD8+ CAR-T cells in the peripheral blood of the mice at the indicated time post CAR-T cell transfer. Two-way ANOVA with Tukey’s multiple comparison test.
Supplementary information
Supplementary Fig. 1
Gating strategy for flow cytometric analysis of peripheral blood from tumour-bearing NSG mice treated with CAR T cells.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Knudsen, N.H., Escobar, G., Korell, F. et al. In vivo CRISPR screens identify modifiers of CAR T cell function in myeloma. Nature (2025). https://doi.org/10.1038/s41586-025-09489-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41586-025-09489-8