Abstract
Holliday junctions (HJs) are branched four-way DNA structures that link recombining chromosomes during double-strand break repair1. Despite posing a risk to chromosome segregation, HJs accumulate during meiotic prophase I as intermediates in the process of crossing-over2,3. Whether HJs have additional regulatory functions remains unclear. Here we establish an experimental system in budding yeast that enables conditional nucleolytic resolution of HJs after the establishment of meiotic chromosome synapsis. We find that HJ resolution triggers complete disassembly of the synaptonemal complex without disrupting the axis–loop organization of chromosomes. Mechanistically, HJs mediate the continued association of ZMM proteins with recombination nodules that form at the axes interface of homologous chromosome pairs. ZMM proteins, in turn, promote polymerization of the synaptonemal complex while simultaneously protecting HJs from processing by non-crossover pathways. Thus, reciprocal feedback between ZMMs, which stabilize HJs, and HJs, which retain ZMM proteins at future crossover sites, maintains chromosome synapsis until HJ-resolving enzymes are activated during exit from prophase I. Notably, by polymerizing and maintaining the synaptonemal complex structure, the HJ–ZMM interplay suppresses de novo double-strand break formation and recombination reinitiation. In doing so, this interplay suppresses the DNA damage response, enabling meiotic progression without unrepaired breaks and supporting crossover assurance.
Similar content being viewed by others
Main
During meiosis, an evolutionarily conserved proteinaceous structure—the synaptonemal complex (SC)—mediates the synapsis of homologous chromosome pairs to regulate genetic exchange and crossing-over4. The SC comprises filamentous lateral elements that organize chromatin into linear arrays of loops together with cohesin complexes, and a central region that connects the two homologue axes5. The central region of the SC, but not the axes, is thought to have liquid-crystalline properties, where weakly bonded proteins can undergo internal rearrangement and exchange6,7,8,9,10. SC assembly takes place gradually, starting in the zygotene stage and eventually connecting the paired homologue axes along their entire length during the pachytene stage, a process that takes around 2.5 h in budding yeast and several days in mice4,11. How SC formation is initiated and how the SC structure is subsequently maintained to ensure the stable end-to-end engagement of all homologue pairs remain subjects of intense research.
In several organisms, including budding yeast, SC assembly is functionally linked to the repair of developmentally programmed DNA double-strand breaks (DSBs) through homologous recombination12. SC polymerization initiates at a subset of recombination sites where interhomologue DNA joint molecules are progressively stabilized and mature into intermediates containing two four-armed HJs (double HJs, dHJs)2,11,13,14,15. A link between synapsis and recombination is provided by a group of meiosis-specific proteins, collectively known as ZMMs (Zip1–4, Msh4–5, Mer3, Spo16), which bind to and stabilize nascent repair intermediates and coordinate dHJ maturation with SC assembly14,15,16,17,18,19,20,21,22,23. By promoting SC assembly between homologous chromosomes, ZMMs contribute to the downregulation of DSB formation24,25,26,27,28,29. Whether ZMMs are continuously required for the stability of dHJs, for the maintenance of chromosome synapsis and, as such, for suppressing the formation of new DSBs is unclear.
Whereas SC assembly takes place gradually, SC disassembly is linked to a sharp cell cycle transition that drives exit from prophase I and entry into the first meiotic division30. Notably, SC disassembly and dHJ resolution are temporally coordinated by Ndt80-mediated expression of polo kinases, which activate HJ resolvases while also driving the disassembly of SC components31,32,33. Such a tight spatiotemporal relationship between the stabilization and resolution of recombination intermediates and the assembly and disassembly of the SC led us to examine whether recombination intermediates have a previously unknown role in the maintenance of chromosome synapsis. We identified a reciprocal functional interplay between dHJs and ZMM proteins that is crucial for maintenance of the SC structure. We propose that, by supporting chromosome synapsis and, thereby, contributing to the suppression of de novo DSB formation, this dHJ–ZMM interplay coordinates meiotic progression with crossover assurance.
dHJs stabilize the SC during pachytene
To assess whether recombination intermediates are required for the maintenance of chromosome synapsis, we used ndt80-deletion (ndt80∆) mutants in combination with conditional expression of an engineered structure-selective endonuclease, Yen1ON, which can process a broad range of branched DNA repair intermediates34 (Fig. 1a). ndt80∆ mutants arrest meiotic progression with synapsed homologues and unresolved dHJs3,30. Conversely, ectopic expression of Yen1ON is sufficient to drive nucleolytic resolution of HJ-containing recombination intermediates during prophase I35. Yen1ON expression was initiated by the addition of β-oestradiol to cultures 7 h after induction of meiosis, at which point more than 85% of cells displayed fully synapsed chromosomes as assessed by immunostaining the transverse filament protein Zip1 (ref. 36) on chromosome spreads (Fig. 1b,c (7 h in sporulation medium, SPM) and Extended Data Fig. 1a). Notably, expression of Yen1ON resulted in a progressive loss of chromosome-associated Zip1 (Fig. 1b,c). This was accompanied by a sharp increase in nuclei with Zip1 aggregates, known as polycomplexes, which are known to form as a consequence of impaired SC assembly36 (Fig. 1b (white arrowheads) and 1c (dashed line)). Despite the altered localization, Zip1 protein levels were unaffected by Yen1ON expression, suggesting that the loss of Zip1 from chromosomes occurred independently of changes in protein expression or stability (Extended Data Fig. 1b,c). Supporting a causal link between the nucleolytic resolution of recombination intermediates and chromosome synapsis defects, the increase in Yen1ON protein levels mirrored the decreasing proportion of nuclei with fully synapsed chromosomes (Fig. 1d). Moreover, Zip1 loading onto chromosomes remained unchanged in cells expressing a nuclease-deficient Yen1ON variant35, Yen1ON-ND (Fig. 1e–g and Extended Data Fig. 1d,e), or in control cultures in which Yen1ON or Yen1ON-ND were not induced (Extended Data Fig. 1f,g). Similar findings were obtained by following SC dynamics in living cells using Zip1GFP, where expression of Yen1ON, but not Yen1ON-ND, promoted the stepwise loss of chromosome synapsis (Extended Data Fig. 1h–k and Supplementary Videos 1 and 2).
a, The experimental set-up for conditional nucleolytic resolution of HJs after meiotic chromosome synapsis is established. b, Representative images of meiotic chromosome spreads at the indicated times in SPM, immunostained for Zip1 (green); DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; grey). Yen1ON was induced by addition of β-oestradiol (or methanol (MeOH) as a control) at 7 h. The insets show a magnification of the region indicated by a dashed box; Rec8 (magenta) marks chromosome axes. The arrowheads indicate Zip1 polycomplexes. c, Quantification of Zip1 synapsis and polycomplexes from b. n = 50 nuclei per timepoint, representative of three biological replicates. d, Quantification of Yen1ON protein levels from Extended Data Fig. 1b and full Zip1 synapsis from c, normalized to peak values. e, Representative images of meiotic chromosome spreads at the indicated times in SPM as in b, but for PGAL1-YEN1ON-ND cells. f, Quantification of Zip1 synapsis and polycomplexes (as in c) from the experiment in e. g, Quantification of Yen1ON-ND protein levels (as in d) from the experiment in Extended Data Fig. 1d and full Zip1 synapsis from the experiment in f. h, The experimental set-up to inhibit DSB formation by Rec104FRB nuclear depletion and resolve HJs by Yen1ON expression in pachytene-arrested ndt80∆ cells. i, Southern blot analysis of DSBs and DNA joint molecules (JMs) at the HIS4::LEU2 recombination hotspot from h. Gel source and biological replicate data are provided in Extended Data Fig. 3c. j, Quantification of DSBs and joint molecules from the experiment in i and a biological replicate, with or without Rec104FRB nuclear depletion. Data are the mean and range of the percentage of total DNA. k, Quantification of Zip1 synapsis and polycomplex formation from the experiment in i, with or without Rec104FRB nuclear depletion, including an additional timepoint (11 h in SPM). n = 50 nuclei per timepoint, representative of two biological replicates. l, Quantification of joint molecules as in j, after Rec104FRB nuclear depletion and with or without Yen1ON induction. m, Quantification of Zip1 synapsis and polycomplex formation as in k, after Rec104FRB nuclear depletion and Yen1ON induction. n = 50 nuclei per timepoint, representative of two biological replicates. For b and e, scale bars, 2 µm.
We next expanded our analysis to other components of the SC. Immunostaining of the central element protein complex Ecm11–Gmc2 (refs. 37,38), as well as SC-associated Smt3 (also known as SUMO)38, revealed analogous outcomes to Zip1, with progressive loss of chromosome association and accumulation into large aggregates linked to Yen1ON expression (Extended Data Fig. 2a–c). In budding yeast, disassembly of the SC central element during pachytene exit coincides with the loss of chromosome axis components and partial release of Rec8-containing cohesin, a process driven by Ndt80-dependent expression of polo kinase Cdc5 (refs. 31,32,39). After Yen1ON expression, Rec8 remained localized to chromatin, retaining a linear pattern of accumulation even in regions in which chromosome synapsis was lost (Extended Data Fig. 2d,e). However, we did notice that a substantial fraction of nuclei exhibited partially disorganized Rec8 threads, possibly due to the local separation of homologous chromosomes (Extended Data Fig. 2e (light pink)). In agreement with this view, super-resolution stimulated emission depletion (STED) microscopy revealed frequent chromosome axis splitting, which correlated with Zip1 loss (Extended Data Fig. 2f). These findings suggest that resolution of recombination intermediates leads to the loss of chromosome synapsis without significantly disrupting the axis–loop organization of meiotic prophase I chromosomes.
Meiotic DSBs continue to form in pachytene-arrested ndt80∆ mutants, albeit at low levels compared with peak DSB formation during the leptotene stage3,25,30 (see also Fig. 1i,j, with data at the HIS4::LEU2 recombination hotspot13). Thus, downstream formation of nascent recombination intermediates could have a role in maintaining the SC structure. To examine this possibility, we performed conditional nuclear depletion of Rec104, an essential factor in meiotic DSB formation, using the rapamycin-dependent anchor-away system40 (Fig. 1h). Addition of rapamycin to the pachytene-arrested cultures strongly reduced DSB levels and halted the accumulation of DNA joint molecules (Fig. 1i,j (light red) and Extended Data Fig. 3a–c). Moreover, analysis of DNA joint molecules in two-dimensional gels showed stabilization of four-armed recombination intermediates containing dHJs, as well as three-armed single-end intermediates that are likewise crossover-designated2,13 (Extended Data Fig. 3d,e (light red)). Importantly, inhibition of DSB formation did not severely interfere with chromosome synapsis, even though we observed that a slightly higher proportion of nuclei contained partial Zip1 synapsis (Fig. 1k (light green)). Expression of Yen1ON after DSB inhibition by Rec104FRB depletion resulted in the resolution of all remaining recombination intermediates at HIS4::LEU2 and a complete loss of Zip1 from chromosomes (Fig. 1l,m (dark red) and Extended Data Fig. 3d–g (dark red)). These data suggest that DSB formation, although initially required for SC assembly, is largely dispensable for SC maintenance. Instead, SC maintenance relies on long-lived recombination intermediates, most likely containing dHJs.
dHJ–ZMM interplay maintains the SC
As some ZMM proteins are known to preferentially bind to HJ DNA in vitro, and all localize to future crossover sites in vivo15,16,17,18,19,20,21,23, we posited that Yen1ON-mediated cleavage of dHJs could lead to loss of ZMMs from chromosomes. In agreement, we observed a significant loss of chromosome-associated Zip3, Msh5 and Zip4 foci, which accumulated in polycomplexes after Yen1ON expression (Fig. 2a,b and Extended Data Fig. 4a–e). The three selected ZMMs represent functional subgroups: Msh5 forms with Msh4 the MutSγ complex, Zip4 forms with Zip2 and Spo16 a functional ZMM subcomplex called ZZS, and Zip3 is an E3 ligase23. It is therefore likely that all ZMM proteins directly or indirectly require recombination intermediates for continued chromosome association during pachytene.
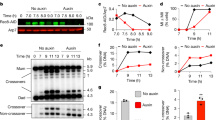
a, Representative chromosome spreads at the indicated times in SPM, after Yen1ON induction by β-oestradiol addition (or methanol control) at 7 h, immunostained for Ecm11–Gmc2 (green) and Zip3 (magenta). The arrowheads mark Zip3 localizing to the Ecm11–Gmc2 polycomplex. b, Quantification of Zip3 focus number from a. Data are mean ± s.d. n = 30 nuclei per timepoint. Statistical analysis was performed using Kruskal–Wallis tests with Dunn’s multiple-comparison test (P ≤ 0.001). Representative of two biological replicates. c, The experimental set-up for conditional depletion of a ZMM protein in pachytene-arrested ndt80∆ cells. Os, Oryza sativa. d, Western blot analysis of Zip3AID levels in cells at the indicated times in SPM, treated as described in c. Crm1 was used as the protein loading control. The asterisks indicate putative SUMOylated Zip3. Representative of two biological replicates. e, Representative images of chromosome spreads from d, immunostained for Zip1 (green) and Zip3AID (magenta). ZIP3AID cells exhibit frequent polycomplex formation but show normal spore viability (Supplementary Table 2). f, Quantification of Zip1 synapsis and polycomplexes from e. n = 50 nuclei per timepoint, representative of two biological replicates. g,h, Southern blot (g) and quantification of joint molecules and non-crossover (NCO1) and crossover (CO2) products (h) at the HIS4::LEU2 recombination hotspot from d. mcJM, multichromatid DNA joint molecule; P1, parental 1; P2, parental 2. i, The experimental set-up for conditional depletion of a ZMM protein and Sgs1 in pachytene-arrested ndt80∆ cells. j, Western blot analysis of Zip3AID and Sgs1AID levels as in d for ZIP3AIDSGS1AID cells treated as described in i. Representative of two biological replicates. k,l, Southern blot (k) and quantification of joint molecules and non-crossover and crossover products (l) as in g and h, respectively, for the experiment in j. m, Representative images of chromosome spreads as in e, for the experiment in j. Both Zip3AID and Sgs1AID carry the Myc epitope tag. n, Quantification of Zip1 synapsis and polycomplexes as in f, for the experiment in m. n = 50 nuclei per timepoint, representative of two biological replicates. For a, e and m, scale bars, 2 μm.
We next sought to test whether ZMMs are continuously required for the maintenance of chromosome synapsis. To this end, we generated auxin-inducible degron (AID) alleles of ZIP3, MSH4 and ZIP4 (Fig. 2c). Addition of 5-Ph-IAA to pachytene-arrested cultures triggered rapid degradation of Zip3AID, Msh4AID and Zip4AID, resulting in their efficient depletion from chromosomes (Fig. 2d,e and Extended Data Fig. 4f–i). Notably, this resulted in a complete loss of chromatin-associated Zip1, as well as a more disorganized pattern of Rec8 accumulation, indicative of synapsis defects and homologue separation (Fig. 2e,f and Extended Data Fig. 4h–o). These results show that, in addition to their previously described roles in promoting SC assembly, ZMMs are continuously required for SC maintenance.
ZMMs prevent dHJ dissolution by STR
During meiosis, ZMMs have been shown to protect nascent recombination intermediates from disassembly by the Sgs1–Top3–Rmi1 (STR) complex41, but it is unclear whether their function is similarly required to suppress STR-mediated dHJ dissolution42. Physical analysis of recombination revealed a rapid loss of all DNA joint molecules at the HIS4::LEU2 recombination hotspot after Zip3AID or Msh4AID depletion (Fig. 2g,h and Extended Data Fig. 5a–c). Consistent with STR-mediated dHJ dissolution, depletion of Zip3AID or Msh4AID resulted in a specific increase in non-crossover products (Fig. 2h and Extended Data Fig. 5c), and simultaneous depletion of Zip3AID or Msh4AID and Sgs1AID resulted in the stabilization and accumulation of DNA joint molecules (Fig. 2i–l and Extended Data Fig. 5d–f). Note that depletion of Sgs1AID also led to a small reduction in the accumulation of crossovers in ndt80∆ mutants (Extended Data Fig. 5g–i). This observation is consistent with previous research showing that ndt80∆ mutants accumulate a small proportion of Sgs1-dependent crossovers3,43,44. The precise origin of these crossovers remains unclear. Overall, our findings demonstrate that ZMMs are not only required for the formation of crossover-designated recombination intermediates but are also crucial for their continued protection from dissolution into non-crossovers by the STR complex.
To determine whether recombination intermediates contribute to SC maintenance independently of ZMMs, we monitored Zip1 after combined depletion of Zip3AID and Sgs1AID. Cells depleted of Sgs1AID and Zip3AID showed loss of Zip1 from chromosomes, but with a significant delay compared to Zip3AID depletion alone (Fig. 2m,n; compare with Fig. 2e,f). Similar results were obtained by combining conditional depletion of Msh4AID and Sgs1AID (Extended Data Fig. 5j,k). Furthermore, live-cell imaging confirmed these observations, with the structured Zip1GFP signal persisting significantly longer after combined depletion of Zip3AID and Sgs1AID (Extended Data Fig. 5l–n and Supplementary Videos 3 and 4). We then hypothesized that the delayed loss of chromosome synapsis might be due to partial retention of the ZMM subcomplex ZZS on chromosomes, as it can directly bind to recombination intermediates20,21 and has been directly implicated in synapsis initiation22. In support of this model, combined depletion of Zip4AID and Sgs1AID resulted in the rapid loss of Zip1 without delay (Extended Data Fig. 5o–q; compare with Extended Data Fig. 5j,k).
Taken together, these observations suggest that dHJs are required for the continuous association of ZMMs with chromosomes. In turn, ZMMs prevent dHJ dissolution by non-crossover pathways, while also promoting SC maintenance.
dHJs enable reversible SC disassembly
In budding yeast, paired chromosome axes retain the ability to assemble the SC structure even in late prophase I6. To test whether dHJs function to stabilize sites of synapsis initiation to enable continuous reinitiation of SC polymerization and maintenance of the SC structure, we developed a molecular tool to reversibly disassemble the SC without loss of dHJs (Fig. 3a). Conceptually, this tool takes advantage of Smt3 constituting a reversible post-translational protein modifier with essential roles in SC assembly37,38,45 as well as in dHJ processing46,47. In the first of two steps, we examined whether conditional protein deSUMOylation is sufficient to disassemble the SC in pachytene-arrested cells. To address this, we generated ndt80∆ strains conditionally expressing a truncated version of the Smt3 isopeptidase Ulp1 (ref. 48) (Ulp1∆N; Extended Data Fig. 6a). In contrast to Ulp1, Ulp1∆N is unable to associate with the nuclear pore complex, thereby extending its activity beyond the nuclear periphery. In addition to placing ulp1∆N under an β-oestradiol-inducible promoter, we also fused ulp1∆N to the FKBP12–rapamycin-binding (FRB) domain (ulp1∆N-FRB), which was necessary in a subsequent second step to enable nuclear depletion using the anchor-away system (Extended Data Fig. 6b). Induction of Ulp1∆N-FRB expression using β-oestradiol resulted in a rapid decrease in global Smt3-modified protein levels, including polySUMOylated forms of Ecm11 (Extended Data Fig. 6c)—one of the most prominent substrates for polySUMOylation during budding yeast meiosis37,45. As a result, we observed complete loss of Smt3 and Zip1 from chromosomes within 1 h of Ulp1∆N-FRB induction (Fig. 3b,c and Extended Data Fig. 6d). Central to our approach, we found that expression of Ulp1∆N did not alter the level of DNA joint molecules at HIS4::LEU2 (Fig. 3d,e and Extended Data Fig. 6e) and the co-alignment of homologue axes was largely maintained (Extended Data Fig. 6f,g). In a second step, we tested whether the subsequent nuclear depletion of Ulp1∆N-FRB would enable the re-establishment of the SC. Notably, addition of rapamycin to the cultures resulted in the recovery of protein SUMOylation and reassembly of Zip1 along chromosomes within 30 min (9.5 h in SPM), and complete restoration of the SC structure within 1 h (10 h in SPM) (Fig. 3b,c and Extended Data Fig. 6c,d). Ulp1∆N-FRB expression did not noticeably alter DSB levels (Extended Data Fig. 6e), and SC reassembly did not depend on newly arising DSBs, as experimentally confirmed by monitoring Zip1 synapsis after Rec104AID depletion (Extended Data Fig. 6h–j).
a, The experimental set-up for reversible SC disassembly in pachytene-arrested ndt80∆ cells through conditional protein deSUMOylation using the Smt3 isopeptidase mutant ulp1∆N-FRB. b, Representative images of meiotic chromosome spreads from the experiment in a at the indicated times in SPM, immunostained for Zip1 (green) and Smt3 (magenta). c, Quantification of Zip1 synapsis from b. n = 50 nuclei per timepoint, representative of two biological replicates. d,e, Southern blot (d) and quantification of joint molecules (e) at the HIS4::LEU2 recombination hotspot. Ulp1∆N was induced by β-oestradiol (β-oest.) addition (or methanol as a control) at 7 h in SPM. Data are the mean and range of two biological replicates. Gel source and biological replicate data are provided in Extended Data Fig. 6e. f, The experimental set-up to investigate whether dHJs are required for SC reassembly. g, Representative images of meiotic chromosome spreads from the experiment in f at the indicated times in SPM, immunostained for Zip1 (green). h, Quantification of Zip1 synapsis from g (n = 50 nuclei per timepoint). i, Time-lapse image montage of Zip1GFP in a cell nucleus after β-oestradiol-induced Ulp1∆N-FRB expression (t = 0 min; ~7 h in SPM) and rapamycin-induced nuclear depletion (t = 120 min). Corresponds to Supplementary Video 6. j, Quantification of structured Zip1GFP signal from i. The arrow and dashed line indicate the timepoint of rapamycin addition. Data are the mean of two biological replicates. n = 40 cells each. k, Time-lapse image montage as in i, for the simultaneous expression of Ulp1∆N-FRB and Yen1ON by β-oestradiol addition. Corresponds to Supplementary Video 7. l, Quantification of structured Zip1GFP signal as in j, for the experiment in k. Data are the mean of two biological replicates. n = 40 cells each. Scale bars, 2 μm (b and g) and 1 μm (i and k).
To experimentally test whether dHJs are required for SC reassembly, Yen1ON was expressed simultaneously with Ulp1∆N-FRB (Fig. 3f). Under these conditions, most nuclei failed to re-establish full Zip1 synapsis, even 4 h after nuclear depletion of Ulp1∆N-FRB (12 h in SPM) (Fig. 3g,h and Extended Data Fig. 7a–c). We further confirmed these findings by visualizing SC disassembly and reassembly in single cells. Live-cell imaging of Zip1GFP showed reversible SC disassembly by Ulp1∆N-FRB expression/nuclear depletion in all analysed cells (Fig. 3i,j, Extended Data Fig. 7d,e and Supplementary Videos 5 and 6). Moreover, SC reassembly was severely disrupted when Yen1ON was co-expressed with Ulp1∆N-FRB (Fig. 3k,l and Supplementary Video 7). Overall, these data support a model in which dHJs are sufficient to maintain interhomologue connections in the absence of the central region of the SC. These connections enable the complete re-establishment of chromosome synapsis, presumably by providing a platform that positions ZMM proteins at the axis interface of homologue pairs. In support of this interpretation, we found that, while many nuclei contained Zip3 aggregates after Ulp1∆N-FRB expression, the number of Zip3 foci remained largely unchanged (Extended Data Fig. 7f–h). Furthermore, Yen1ON expression resulted in a marked reduction in the number of Zip3 foci (Extended Data Fig. 7i–k), indicating that Zip3 associates with dHJs and that its retention—and, by extension, the retention of other ZMMs—underlies the rapid SC reassembly after Ulp1∆N-FRB depletion.
dHJ–ZMM feedback limits DSB formation
In organisms in which DSB formation is required for chromosome synapsis, such as budding yeast and mice, SC assembly is thought to suppress the formation of new DSBs on chromosomes that have already successfully engaged in crossover repair24,25,26,27,28,29. This feedback control involves the displacement of HORMAD proteins from chromosome axes, leading to the loss of a DSB-competent state24,26. We therefore hypothesized that SC maintenance through the functional interplay between dHJs and ZMMs might be important for continued downregulation of DSB formation. Indeed, Yen1ON expression in pachytene-arrested cultures led to reaccumulation of Hop1 (also known as HORMAD) on meiotic chromosomes that had lost Zip1 (Fig. 4a,b). This was accompanied by increased Hop1 phosphorylation at Thr318 by the DNA-damage response kinases Mec1 (ATR) and Tel1 (ATM), which occurs in response to meiotic DSB formation49 (Fig. 4c). We confirmed this inference by directly monitoring DSBs at two recombination hotspots (CCT6 and ERG1) using Southern blotting, with DSB levels increasing by around 3–4-fold after Yen1ON induction (Fig. 4d,e and Extended Data Fig. 8a–d). We predicted that the loss of chromosome synapsis after conditional ZMM depletion during pachytene should also result in the re-establishment of a competent state for DSB formation. Indeed, we observed an increase in Hop1 Thr318 phosphorylation after auxin-mediated depletion of Zip3AID, Msh4AID and Zip4AID, and an approximately 3–6-fold increase in DSB levels after Msh4AID depletion (Fig. 4f–h and Extended Data Fig. 8e–h). These findings suggest that dHJs and ZMM proteins have a key role in maintaining a suppressive state for DSB formation by contributing to the maintenance of chromosome synapsis.
a, Representative images of chromosome spreads at peak DSB formation (4 h), and before (7 h) and after (12 h) Yen1ON induction by addition of β-oestradiol (or methanol control), immunostained for Zip1 (green) and Hop1 (magenta). b, The total Hop1 signal intensity per nucleus from a. Data are mean ± s.d. n = 50 nuclei per timepoint. Statistical analysis was performed using Kruskal–Wallis tests with Dunn’s multiple-comparison test (P ≤ 0.001). c, Western blot analysis of the levels of Hop1 and Hop1 phosphorylated at Thr318 from a. p-Hop1, phosphorylated Hop1. Pgk1 was used as the protein loading control. d,e, Southern blot analysis of DSBs at the CCT6 locus (d) and quantification at CCT6 and ERG1 (e). Yen1ON was induced by β-oestradiol addition (or methanol control) at 7 h in SPM. Sae2AID was simultaneously depleted by addition of 5-Ph-IAA (or DMSO control) to prevent DSB repair. Data are the mean and range of fold changes relative to the Sae2AID depletion control from two biological replicates. Gel source and replicate data are provided in Extended Data Fig. 8c. P, parental. f, Western blot analysis as in c for Msh4AID depletion at 7 h in SPM. Representative of two biological replicates. g,h, Southern blot analysis of DSBs (g) and quantification of CCT6 and ERG1 (h), as in d and e, for the experiment in f. Gel source and replicate data are provided in Extended Data Fig. 8e. i, Time-lapse montage of Zip1GFP and Nup84mCherry after Yen1ON induction in early/mid-zygotene (t = 0 min, ~5 h in SPM). The arrowheads mark Zip1GFP aggregates. Nup84mCherry brightness was adjusted for signal visibility. Corresponds to Supplementary Video 9. j–l, Quantification of structured Zip1GFP signal (j), meiosis I (k) and sporulation (l) from the experiment in i. The arrow and dashed line indicate β-oestradiol addition. PC, polycomplex. Data are the mean and range (error bars) of two biological replicates, n = 20 cells each. Scale bars, 2 μm (a) and 1 μm (i).
Premature loss of dHJs impairs meiosis I
To determine whether recombination intermediates are required for SC maintenance during meiotic progression in wild-type (NDT80) cells, we combined timed Yen1ON expression with live-cell imaging of Zip1GFP. We chose to initiate Yen1ON expression around 5 h after induction of meiosis as DNA replication was largely completed (Extended Data Fig. 9a) and cells in different substages of prophase I could be identified on the basis of the Zip1GFP signal: pre-leptotene/leptotene cells lacking structured Zip1GFP; early zygotene cells with a partially structured Zip1GFP signal; and late zygotene/pachytene cells with long Zip1GFP threads throughout the nucleus. Under our imaging conditions, zygotene lasted about 55 min, pachytene around 115 min and anaphase I was initiated around 45 min after exit from pachytene (Extended Data Fig. 9b and Supplementary Video 8). As robust accumulation of Yen1ON requires 45–60 min (Extended Data Fig. 9c,d), and significant processing of DNA joint molecules by Yen1ON becomes detectable only after 1–2 h (Extended Data Fig. 3f,g), we decided to follow the subpopulation initially in early to mid-zygotene at the time of Yen1ON induction to determine the impact of Yen1ON specifically during pachytene. As predicted, cells induced to express Yen1ON in early to mid-zygotene successfully entered pachytene and reached full chromosome synapsis (Fig. 4i,j and Supplementary Video 9). However, 90% of cells reverted from full chromosome synapsis to a state of partial synapsis, with conspicuous Zip1GFP aggregates (Fig. 4i,j). Notably, we also found that cells that reverted to a state of partial synapsis showed a long delay in completing prophase I and eventually failed to undergo the first meiotic division and form spores (Fig. 4j–l and Supplementary Video 9). By contrast, control methanol-treated cultures disassembled the SC, completed meiosis I nuclear division and formed spores efficiently (Fig. 4k,l, Extended Data Fig. 9e,f and Supplementary Video 10). Importantly, Yen1ON expression did not interfere with SC assembly and disassembly, nor with nuclear division, in cells that were in pachytene at the time of induction (Extended Data Fig. 9g–i and Supplementary Video 11). However, Yen1ON expression prevented almost all pre-leptotene/leptotene cells from reaching a state of full synapsis and completely blocked meiosis I nuclear division (Extended Data Fig. 9j–l and Supplementary Video 12). To confirm these findings, we induced Yen1ON expression from the strong copper-inducible PCUP1 promoter in pachytene-arrested ndt80∆ mutants and subsequently triggered pachytene exit by expressing Ndt80 from the PGAL1 promoter. Cells expressing Yen1ON before Ndt80 induction disassembled Zip1GFP but retained a large polycomplex for a prolonged time (Extended Data Fig. 9m–p and Supplementary Videos 13 and 14). Eventually, the Zip1GFP signal was lost and cells attempted to undergo meiosis I nuclear division, as indicated by HTB1mCherry labelling of chromatin. However, most failed, while those that underwent anaphase I exhibited DNA bridges or aberrant nuclear mass distribution (Extended Data Fig. 9q). Overall, these findings suggest that premature processing of recombination intermediates during wild-type meiosis leads to disruption of chromosome synapsis. As a consequence, cell cycle progression is disrupted, most likely due to the formation of de novo DSBs and their repair intermediates, which trigger DNA damage signalling50,51, as observed after Yen1ON expression or ZMM depletion in ndt80∆ mutants (Fig. 4a–h and Extended Data Fig. 8).
dHJ resolution promotes SC disassembly
SC disassembly during exit from pachytene is temporally coordinated with HJ resolution through Ndt80-mediated expression of polo kinase Cdc5 (also known as PLK)31,32. However, it is unclear whether HJ processing contributes to SC disassembly. To test this possibility, we analysed meiotic cells lacking all four HJ resolvases: mlh3∆ mms4mn slx1∆ yen1∆, hereafter, quadruple-resolvase mutant. First, we confirmed that ectopic expression of Cdc5 was sufficient to induce Zip1 loss from the chromosomes of pachytene-arrested ndt80∆ cells, as previously reported32 (Fig. 5a,b (left) and Extended Data Fig. 10a (left)). Notably, quadruple-resolvase mutants retained chromosome-associated Zip1 for a prolonged period (Fig. 5a,b (right) and Extended Data Fig. 10a (right)), despite a significant reduction in Zip1 protein levels as observed in the control cells (Extended Data Fig. 10b,c). Live-cell imaging of Zip1GFP confirmed the delayed loss of chromosome synapsis in quadruple-resolvase mutant cells after Cdc5 expression (Fig. 5c–e and Supplementary Videos 15 and 16). As DNA joint molecules in the quadruple-resolvase mutant may be processed alternatively by the STR pathway, we combined a meiotic-null allele of SGS1 (sgs1mn)41 with the quadruple-resolvase mutant. In this quintuple mutant, in which DNA joint molecules remain unprocessed and accumulate to high levels44, SC disassembly was further delayed compared with the control and quadruple-resolvase mutant but still completed in about 50% of cells within the experimental timeframe (Extended Data Fig. 10d–j and Supplementary Video 17). Next, we examined whether HJ processing contributes to SC disassembly at the prophase-I-to-metaphase-I transition in NDT80 cells. Notably, SC disassembly was delayed relative to spindle pole body (SPB) separation in both the quadruple and the quintuple mutants (Fig. 5f). Nonetheless, the Zip1GFP signal was eventually lost in almost all nuclei analysed (Fig. 5f). The SC disassembly delay was confirmed by the analysis of the Zip1 signal relative to SPB separation on chromosome spreads. At the onset of metaphase I, around 57% of quadruple-resolvase mutants retained Zip1 stretches compared with around 28% of controls (Extended Data Fig. 10k). This effect was more prominent in quintuple mutants (~79%), with approximately 55% still showing Zip1 stretches in nuclei with fully separated SPBs. In summary, these findings suggest that Cdc5 promotes SC disassembly through two distinct mechanisms. One is linked to the processing of HJs, which disrupts SC maintenance, whereas the other probably involves Cdc5-mediated phosphorylation of SC components leading to their degradation, as previously suggested52,53.
a, Representative chromosome spreads at the indicated times in SPM, with immunostaining for Zip1 (green). Cdc5 was induced by β-oestradiol addition at 7 h. b, Quantification of Zip1 synapsis and polycomplex formation from a. n = 50 nuclei per timepoint. c,d, Time-lapse montage of Zip1GFP (c) and quantification of structured Zip1GFP (d) in cells of the indicated genotypes after Cdc5 induction by β-oestradiol addition at around 7 h in SPM (t = 0 min). n = 40 cells. See Supplementary Videos 15 and 16. e, The time to complete loss of structured Zip1GFP signal from d. Data are mean ± s.d. n = 40 cells per genotype. Statistical analysis was performed using two-tailed unpaired Mann–Whitney U-tests. f, Time-lapse montage of Zip1GFP and Cnm67tdTomato in cells of the indicated genotypes (left) and quantification of Zip1GFP signal loss relative to SPB separation (right). Data are the median (dashed line), and first and third quartiles (dotted lines). n = 60 cells per genotype. Statistical analysis was performed using Kruskal–Wallis tests with Dunn’s multiple-comparison test (P ≤ 0.001). g,h, dHJ–ZMM protein interplay coordinates meiotic progression with crossover assurance. g, ZMMs promote the formation of dHJs, which maintain ZMM association at homologue axis interfaces. In turn, ZMMs protect dHJs from STR-mediated dissolution and promote SC assembly and maintenance. MutLγ resolves dHJs into crossovers, contributing to ZMM displacement and loss of SC maintenance. h, SC assembly induces chromosome-autonomous and, eventually, nucleus-wide DSB downregulation. DSB repair completion silences the DNA damage response, enabling Ndt80-mediated pachytene exit without unrepaired DSBs and at least one dHJ per homologue pair. Ndt80-induced Cdc5 activates HJ resolvases, displacing ZMMs and destabilizing the SC to promote rapid SC disassembly. Scale bars, 2 μm (a) and 1 μm (c and f).
Discussion
Our study and the accompanying work by Tang et al.54 support a model in which reciprocal feedback between recombination intermediates and ZMM proteins stabilizes both DNA-based and protein-based interhomologue connections throughout the extended meiotic prophase I (Fig. 5g). Mechanistically, ZMM proteins protect a subset of recombination intermediates from STR-mediated dissolution into non-crossovers. Conversely, these long-lived recombination intermediates function as platforms for the continued association of ZMM proteins with chromatin. Importantly, by stabilizing and precisely positioning ZMMs at the axes interface of recombining homologue pairs, recombination intermediates have a key role in the establishment and maintenance of chromosome synapsis, a process that we postulate involves continuous polymerization of the SC. Our study also suggests that dHJ–ZMM-mediated SC maintenance contributes to the stable suppression of DSB formation on recombining chromosomes (Fig. 5h). Consistent with this view, premature resolution of recombination intermediates during pachytene triggers SC disassembly, de novo DSB formation and DNA damage signalling, ultimately disrupting entry into the first meiotic division.
More broadly, we propose that the reciprocal interplay between dHJs and ZMM proteins constitutes a regulatory circuit that coordinates exit from prophase I with crossover assurance (Fig. 5h). In this model, dHJ-dependent maintenance of chromosome synapsis provides a straightforward mechanism to couple meiotic cell cycle progression with the accumulation of crossover precursors. Only when dHJs accumulate successfully will the SC stably inhibit de novo DSB formation in a chromosome-autonomous manner. This inhibition is in turn necessary to suppress the DNA damage response throughout the genome, which would otherwise delay meiotic progression as long as DSBs or their repair intermediates are present50,51. dHJs possess a unique property that is central to this model: in contrast to DSBs and early DSB repair intermediates, which typically contain extensive regions of single-stranded DNA, late recombination intermediates containing dHJs are unlikely to activate the DNA damage response. This notion is supported by studies showing that HJ resolvase mutants exit prophase I in the presence of high levels of dHJs and undergo catastrophic chromosome segregation33,43,44. Thus, once ZMM-stabilized dHJs accumulate on all chromosomes and synapsis permanently downregulates DSB formation, cells can proceed to the first meiotic division assured of no unrepaired breaks and at least one crossover precursor per homologue pair. A crossover outcome is further enforced by dHJ-association of specific ZMMs, in particular MutSγ, which is thought to mediate the orientation-specific loading and activation of MutLγ (Mlh1–Mlh3) to resolve dHJs in a crossover-specific manner44,55,56. It is interesting to consider that, by enabling cells to silence the DNA damage response, dHJs promote the cell cycle transition that leads to the activation of HJ resolvases, triggering their own turnover. For these reasons, we propose that dHJs are more than passive intermediate structures that form in the process of crossing-over. They have a central regulatory function in coordinating meiotic progression with crossover assurance. Our work also demonstrates that HJs can mediate reversible disassembly of the SC. This reversibility may facilitate the resolution of chromosomal interlocks and contribute to the destabilization of interactions between non-allelic sequences.
We envision that related principles may operate in organisms such as plants or mice, in which DSB formation precedes chromosome synapsis12. In other organisms, such as worms and flies, in which SC assembly occurs independently of DSB formation57,58, changes in SC dynamics have been reported to occur in response to the stabilization of crossover precursors7,8,9,59,60,61. Although dHJs may not be required for SC maintenance in these organisms, they may have a similar key regulatory role in coordinating the exit from prophase I with crossover assurance.
Finally, our findings suggest that the resolution of recombination intermediates is part of the SC-disassembly process (Fig. 5h). Although further work is required to dissect the events leading to SC disassembly, we envision that the trigger for this substantial change in chromosome organization involves at least two distinct processes driven by polo kinase Cdc5: (1) dHJ resolution, which destabilizes ZMM association and, as such, eliminates synapsis (re)initiation sites; and (2) phosphorylation-dependent degradation of SC components, which probably destabilizes the SC more broadly52,53.
Methods
Strain construction
Saccharomyces cerevisiae strains used in this study were SK1 derivatives, as described in Supplementary Table 1. The following alleles have been previously described: ndt80Δ62, YEN1ON (ref. 34), YEN1ON-ND (ref. 35), PGAL1-YEN1ON-3FLAG-2TEV-10HIS35, PGPD1-GAL4(848).ER63, HIS4::LEU2 alleles for physical analysis of recombination13, REC104-FRB-3HA40, fpr1∆64, RPL13A-FKBP12 (ref. 64), PCUP1-OsTIR1F74G (refs. 65,66), PCLB2-3HA-MMS4 (ref. 33), PCLB2-3HA-SGS1 (ref. 41), yen1∆35, mlh3∆35, PGAL1-CDC5-3HA62, PGAL1-NDT80 (ref. 63) CNM67-tdTomato62, HTB1-mCherry62 and ZIP1::GFP700 (marked with HphMX4 170 bp after the stop codon)62,67. YEN1ON-ND is a nuclease-dead version of YEN1ON (ref. 35), carrying the E193A and E195A mutations34. To generate PGAL1-YEN1ON and PGAL1-YEN1ON-ND, the endogenous promoter of C-terminally 18myc-tagged YEN1ON or YEN1ON-ND35 was replaced with the PGAL1 cassette, PCR-amplified from plasmid pYM-N22 (ref. 68). Similarly, PCUP1-YEN1ON was constructed by replacing the endogenous promoter of C-terminally 18myc-tagged YEN1ON with the PCUP1 cassette, PCR-amplified from plasmid pYM-N1 (ref. 68). Nup84 was C-terminally tagged with mCherry (NUP84mCherry) by integrating the 3xmCherry cassette, PCR-amplified from plasmid pFA6a-3xmCherry-KanMX4 (ref. 69). C-terminal tagging of ZIP3, MSH4, ZIP4, SGS1, REC104 and SAE2 with the AID tag was performed by integration of the AID*−9myc cassette (here referred to as AID; AID* comprises IAA17 (amino acids 71–114)), PCR-amplified from plasmid pHyg-AID*−9myc70. ZIP4 was 9myc-tagged at the C terminus (ZIP49myc) by integration of the 9myc-KITRP1 cassette, PCR-amplified from plasmid pWZV86-9myc-KITRP1 (pWZV87) as described previously69. The functionality of the tagged proteins was validated by tetrad microdissection and assessment of spore viability, as described in Supplementary Table 2. ulp1∆N was first published as ∆Nulp1-GFP, in which part of the nuclear pore complex anchoring domain (amino acids 172 to 340) was replaced by GFP48. C-terminal tagging of ulp1∆N with the FKBP12–rapamycin-binding domain (FRB) was performed by integration of a FRB cassette, PCR-amplified from the plasmid pFA6a-FRB-KanMX6 (ref. 64). ULP1 is an essential gene, and therefore strains carrying β-oestradiol-inducible ulp1∆N or ulp1∆N-FRB (PGAL1-ulp1∆N, PGAL1-ulp1∆N-FRB) were generated by replacing the endogenous promoter in a heterozygous strain with the PGAL1 cassette, PCR-amplified from plasmid pYM-N23 (ref. 68). In meiotic experiments using the rapamycin-dependent anchor-away system64, the TOR1 mutation tor1-1 to enhance rapamycin resistance is omitted from the strains. slx1∆ was generated by replacing the SLX1 coding region with the selectable marker hphMX6, PCR-amplified from plasmid pFA6a-hphMX6 (ref. 71). Further details on strain construction and primer sequences used are available on request.
Meiotic time courses
Meiotic time courses were performed with diploid SK1 strains as previously described72. In brief, cells selected on YP-glycerol plates (20 g l−1 bactopeptone, 10 g l−1 yeast extract, 2% (v/v) glycerol, 20 g l−1 agar) for 48 h at 30 °C were plated in small patches on YPD plates (20 g l−1 bactopeptone, 10 g l−1 yeast extract, 20 g l−1 dextrose, 20 g l−1 agar) and grown for about 24 h at 30 °C. Cells were further expanded on YPD plates to form a lawn covering the entire plate, grown for around 24 h at 30 °C and inoculated into pre-sporulation medium (YPA; 20 g l−1 bactopeptone, 10 g l−1 yeast extract, 2% (w/v) potassium acetate) to an optical density at 600 nm (OD600) of about 0.3. Cells were grown and arrested in G1 for 14 h at 25 °C, washed with prewarmed sporulation medium (SPM; 2% (w/v) potassium acetate) and inoculated into SPM to an OD600 of about 3.5. This time defines 0 h in all meiotic time-course experiments. Large meiotic cultures were prepared after scaling up the above protocol and using a 10 l fermenter system as previously described72,73,74. Progression through pre-meiotic DNA replication was followed by fluorescence-activated cell sorting (FACS) analysis of DNA content. Pachytene arrest in ndt80∆ strains was defined as 7 h after induction of meiosis in SPM, unless otherwise indicated. Expression from PGAL1 through GAL4(848).ER was induced by addition of 2 µM β-oestradiol (dissolved in methanol), except for PGAL1-NDT80, which was induced with 1 µM. Expression of Yen1ON from PCUP1 was induced by the addition of 2 µM CuSO4 (dissolved in H2O). To deplete AID-tagged proteins, PCUP1-OsTIR1F74G was induced by the addition of 50 µM CuSO4 approximately 30 min before the addition of 100 µM 5-Ph-IAA (dissolved in DMSO). Anchor-away experiments were performed by addition of 1 µg ml−1 rapamycin (dissolved in DMSO).
FACS analysis of DNA content
Cellular DNA content was determined to monitor release from G1 arrest and entry into the pre-meiotic S phase. In brief, 1 ml of meiotic culture was collected and fixed in 70% (v/v) ice-cold ethanol. Cells were washed once and resuspended in 50 mM Tris-HCl pH 7.5. RNA was digested for 2–4 h at 37 °C by the addition of 2 µl RNase (100 mg ml−1). Cells were washed once in FACS buffer (200 mM Tris-HCl pH 7.5, 211 mM NaCl, 78 mM MgCl2), resuspended in FACS buffer containing 50 µg ml−1 propidium iodide and sonicated briefly. An aliquot was diluted 10–20 times in 1 ml 50 mM Tris-HCl pH 7.5 and DNA content was measured using the FACSCalibur cytometer (BD Biosciences) controlled by the CellQuest software (BD Biosciences). Cytometer data were analysed using FlowJo software (BD Biosciences).
Meiotic chromosome spreads
Yeast chromosome surface spreads were prepared as described in refs. 75,76, with some modifications. In brief, 1 ml of meiotic culture collected at the indicated timepoint was centrifuged (4 min at 700 rcf) and cells were resuspended in 200 µl spheroplasting solution (2% (w/v) potassium acetate, 0.8 M sorbitol). Cells were incubated with 10 mM DTT for 15 min at 30 °C and then digested with 5 µl Zymolyase 20T solution (10 mg ml−1) for around 10 min. Spheroplasting efficiency was monitored by mixing an aliquot of 2 µl with 2 µl of 2% (w/v) sarcosyl, which should lyse the spheroplasted cells. The digestion was stopped by adding 400 µl of ice-cold stop solution (0.1 M MES, 1 mM EDTA, 0.5 mM MgCl2, 1 M sorbitol, pH 6.4) and the spheroplasts were centrifuged (4 min at 700 rcf) and resuspended in 100 µl of stop solution. Immediately after, 20 μl of spheroplast suspension was processed on a clean glass slide by sequentially adding 40 μl of fixative (4% (w/v) paraformaldehyde, 3.4% (w/v) sucrose) for pre-fixation, 80 μl of 1% (v/v) Lipsol to initiate lysis and 80 μl of fixative for final fixation. The slides were dried overnight in a fume hood before immunostaining.
For immunostaining, slides were washed once for 15 min in 1× PBS and blocked for 20 min with 200 μl blocking buffer (1% (w/v) BSA and 0.2% (w/v) gelatin in 1× PBS). The slides were then incubated with 50–80 μl diluted primary antibodies (in blocking buffer) covered with a coverslip in a humidity chamber for either 4 h at room temperature or overnight at 4 °C. The slides were then washed three times in 1× PBS for 5 min and incubated with diluted secondary antibodies covered with a coverslip in a humidity chamber for 2–4 h at room temperature. Finally, the slides were washed three times in 1× PBS for 5 min and mounted with ProLong Diamond Antifade Mountant (Invitrogen) containing DAPI to visualize DNA.
The following primary antibodies were used: rabbit anti-Zip1 (1:500)77, guinea pig anti-Rec8 (1:1000)78, rabbit anti-Zip3 (1:1,000)79, rabbit anti-Msh5 (1:500)79, mouse anti-Myc (1:300, 9E10, Cancer Research UK), guinea pig anti-Ecm11-Gmc2 (1:800)80, mouse anti-Smt3 (1:500, 4F2.F5.G2, Rockland Immunochemicals), guinea pig anti-Hop1 (1:500)81 and mouse anti-γ-tubulin/Tub4 (1:200, MPI-CBG A81)62. Secondary antibodies raised in goat or donkey and conjugated to Alexa Fluor 488, Alexa Fluor 555 and Alexa Fluor 647 (1:500, Invitrogen) were used for detection. For STED, chromosome spreads were prepared as described above and immunostained using goat anti-rabbit STAR ORANGE and goat anti-guinea pig STAR RED secondary antibodies (1:100, Abberior) for detection and slides mounted with ProLong Glass Antifade Mountant (Invitrogen). Widefield fluorescence microscopy images were acquired on a DeltaVision Ultra epifluorescence microscope (GE Healthcare) with a 100× oil-immersion UPlanSApo objective (1.4 NA, working distance 0.13 mm) and a sCMOS camera controlled by AcquireUltra software (v.1.2.3) running on Linux. STED images were acquired using an Abberior STEDYCON mounted on a Zeiss Axio Imager A2 and a 100× oil-immersion alpha Plan-Apochromat objective (1.46 NA, working distance 0.11 mm) and deconvolved using Huygens Professional (SVI) STED deconvolution software. Images were processed and analysed using Fiji82 and ImageJ macros to aid intensity analysis and focus counting. Unless otherwise noted, single z slice images are shown modified for display using linear brightness and contrast adjustments.
The following criteria were used to assess chromosome synapsis based on Zip1, Ecm11–Gmc2 and Smt3 immunostaining. Using the chromosome axes (Rec8) and chromatin (DAPI) as references, three categories of nuclei were defined: ‘full synapsis’ includes nuclei in which ≥75% of chromosomes show complete, continuous synapsis; ‘partial synapsis’ includes nuclei with short stretches of polymerized Zip1, with ≤75% of the chromosomes synapsed; ‘no synapsis’ includes nuclei with no or few foci of Zip1 staining. Nuclei with extrachromosomal Zip1, Ecm11–Gmc2 or Smt3 polycomplexes were also scored, and polycomplexes were quantified separately by monitoring for absence of chromatin association.
Live-cell imaging
All live imaging experiments used conditioned sporulation medium (filter-sterilized SPM from the respective cultures). In brief, 8-well Lab-Tek II chambered coverglasses (Nunc, 155409) were prepared by coating the bottom of each well with 2 mg ml−1 concanavalin A (dissolved in 1× PBS containing 50 mM CaCl2 and 50 mM MnCl2) and incubating at 30 °C for 10 min. Excess concanavalin A was removed by aspiration and each well was washed twice with conditioned SPM. For imaging, cells were collected from the meiotic cultures at the desired timepoints after meiotic induction in SPM and diluted with conditioned SPM to OD600 = 1.8. Subsequently, 0.1 ml aliquots were added to concanavalin-A-coated wells and the cells were allowed to settle at 30 °C for 3 min. The supernatant was removed and the wells were carefully washed once with conditioned SPM before 200 µl of conditioned SPM was added to each well. Cells were imaged using a DeltaVision Ultra epifluorescence microscope (GE Healthcare) with an environmental chamber heated to 30 °C. For experiments involving the addition of β-oestradiol, 5-Ph-IAA or rapamycin, the required chemicals were diluted to twice the final concentration in 200 µl of conditioned SPO. The diluted mixture was then carefully added directly to the respective wells.
Images were acquired using a 60× oil-immersion UPlanXApo objective (1.42 NA, working distance 0.15 mm) and a sCMOS camera controlled by AcquireUltra software (v.1.2.3) running on Linux. z-stack images (8 sections, 1 µm apart) were acquired every 3, 5 or 10 min for 12–15 h. Images were deconvolved using Huygens Professional (SVI) widefield deconvolution software and maximum intensity z-projected over the range of acquisition in Fiji. Maximum-intensity z-projections are shown in time-lapse montages and videos, modified for presentation using linear brightness and contrast adjustments. Images in videos are additionally smoothed using mean filtering in Fiji and aligned using the HyperStackReg ImageJ plugin83. Montages in videos were generated using the Multi Stack Montage ImageJ plugin (BIOP, EPFL).
In situ immunofluorescence analysis
The efficiency of Rec104FRB nuclear depletion using the anchor-away technique was evaluated by in situ immunostaining according to a previously described protocol84, with some modifications. In brief, 1 ml of meiotic culture was fixed with 3.7% formaldehyde overnight. Cells were washed three times with 0.1 M KPi buffer pH 6.4, once with spheroplasting buffer (0.1 M KPi pH 7.4, 1.2 M sorbitol, 0.5 mM MgCl2) and resuspended in 200 µl spheroplasting buffer. Cells were incubated with 10 mM DTT for 15 min at 30 °C and then digested with 10 µl of Zymolyase 100T solution (1 mg ml−1), the efficiency of which was monitored by cell lysis in the presence of 2% (w/v) sarcosyl. Spheroplasts were washed once with spheroplasting buffer, loaded onto poly-l-lysine-coated microscope slides and fixed in ice-cold methanol for 3 min and in ice-cold acetone for 10 s. Cells were blocked in 1% (w/v) BSA in 1× PBS and then stained with a primary mouse anti-HA.11 antibody (1:200, 16B12, BioLegend) and a secondary Alexa Fluor 488-conjugated anti-mouse antibody (1:300, Invitrogen). Slides were mounted with ProLong Diamond Antifade Mountant (Invitrogen) containing DAPI to visualize DNA. Images were acquired on the Zeiss Axio Imager M2 equipped with a 63× oil-immersion Plan-Apochromat objective (1.4 NA, working distance 0.19 mm) and a CoolSNAP HQ2 camera under the control of Zeiss ZEN blue 3.3, and image analysis was performed using Fiji. Approximately 100 cells were analysed per experiment.
Protein analysis by western blotting
Protein extracts were prepared as previously described62. In brief, 10 ml of meiotic culture was collected by centrifugation (3 min at 800 rcf) and the cells were opened in 10% trichloroacetic acid using glass beads and a FastPrep-24 5G instrument (MP Biomedicals) running three cycles of 40 s (6 m s−1). Protein precipitates were collected by centrifugation at 4 °C (10 min at 800 rcf), resuspended in 2× NuPAGE sample buffer (Invitrogen) supplemented with 200 mM DTT and neutralized with 1 M Tris base at a 2:1 (v/v) ratio. The samples were boiled at 95 °C for 5 min and cleared by centrifugation (10 min at 21,300 rcf). The relative protein concentration was measured using the Bio-Rad protein assay. Protein samples were separated on NuPAGE 3–8% Tris-Acetate gels or 4–12% Bis-Tris gels (Invitrogen) using NuPAGE Tris-Acetate or MES SDS running buffer (Invitrogen) and transferred onto Amersham Hybond 0.45 µm PVDF membranes (Sigma-Aldrich).
The following primary antibodies were used for immunoblotting: rabbit anti-Myc conjugated to HRP (1:15,000, ab1326, Abcam), rabbit anti-Zip1 (1:5,000)77, rabbit anti-Crm1 (1:5,000, a gift from K. Weis), mouse anti-Myc (1:5,000, 9E10, Cancer Research UK), mouse anti-GFP (1:2,000, 7.1/13.1, Roche), rabbit anti-Ecm11 (1:5,000, a gift from A. Pichler), rabbit anti-Smt3 (1:5,000, a gift from A. Pichler), mouse anti-Pgk1 (1:10,000, 22C5D8, Invitrogen), guinea pig anti-Hop1 (1:5,000)81, rabbit anti-Hop1-pT318 (1:5,000)81 and mouse anti-HA.11 (1:2,500, 16B12, BioLegend).
The following secondary antibodies were used in Extended Data Figs. 6c and 7b,d: goat anti-mouse immunoglobulins conjugated to HRP (1:10,000, P0447, Agilent) and swine anti-rabbit immunoglobulins conjugated to HRP (1:10,000, P0399, Agilent). For all other experiments, the following fluorescent secondary antibodies were used for detection and quantification of protein levels: goat anti-rabbit conjugated to IRDye 800CW (1:15,000, 926-32211, LI-COR Biosciences), goat anti-mouse IgG conjugated to Alexa Fluor 680 (1:15,000, A21057, Invitrogen) and goat anti-guinea pig IgG conjugated to Alexa Fluor 647 (1:15,000, A21450, Invitrogen). After washing with PBS-T, the blots were imaged using the ChemiDoc MP Imaging System (Bio-Rad) and image analysis and quantification were performed in Fiji. Images were processed using Fiji and Adobe Photoshop, with only linear adjustments to brightness and contrast applied for presentation purposes.
Physical analysis of recombination at HIS4::LEU2 by Southern blotting
Genomic DNA preparation and physical analysis of recombination intermediates at the HIS4::LEU2 recombination hotspot by Southern blotting were performed as previously described2,13,33,85,86. In brief, 50–100 ml of cells was collected from the cultures and resuspended in 0.1 mg ml−1 trioxsalen. DNA was crosslinked at 3,600 mJ cm−2 using a UVP Crosslinker CL-3000L (Analytik Jena), with cells kept on ice and mixed at regular intervals. Genomic DNA was extracted using guanidine/sarcosyl to lyse the cells, followed by phenol–chloroform extraction. After genomic DNA preparation, approximately 1.5 mg of DNA was digested with XhoI (DNA joint molecule analysis) or XhoI/NgoMIV (crossover/non-crossover analysis) and separated by one-dimensional gel electrophoresis on 0.6% agarose gels in 1× TBE buffer (90 mM Tris base, 90 mM boric acid, 2 mM EDTA pH 8.0) at 2 V cm−1 for 21 h. Physical analysis of branched recombination intermediates using native/native two-dimensional gels was performed as previously described87,88 with XhoI-digested genomic DNA loaded onto a 0.4% agarose gel (Seakem Gold, Lonza) in 1× TBE without ethidium bromide and run at 1 V cm−1 for 21 h at room temperature. Gels were stained in 1× TBE containing ethidium bromide and portions of lanes containing DNA species of interest were excised and placed in a gel tray at 90° to the direction of electrophoresis. 0.8% agarose (Ultrapure, Invitrogen) in 1× TBE containing ethidium bromide was poured around the gel slices and allowed to solidify. Two-dimensional gel electrophoresis was performed at 4 °C in pre-chilled 1× TBE containing ethidium bromide at 5.3 V cm−1 for 4 h. After one or two-dimensional gel electrophoresis, the DNA was transferred to a GeneScreen Plus membrane (Revvity) by alkaline transfer. The membranes were hybridized with a probe (Probe A)89 for the HIS4::LEU2 recombination hotspot, which was random-primed labelled with [α−32P]dCTP using High Prime labelling mixture (11585592001, Roche). Hybridized membranes were exposed to a phosphor screen and imaged using the Amersham Typhoon phosphor imager (Cytiva). Southern blot images were adjusted for presentation using Fiji, applying only linear modifications to brightness and contrast. Signal intensities of different recombination intermediates were quantified relative to the total lane signal using ImageQuant software or Fiji. The signal at 0 h in SPM was used for background subtraction.
Physical analysis of DSBs by Southern blotting
Genomic DNA was isolated from cells embedded in low-melting-temperature agarose plugs to prevent non-specific shearing of genomic DNA as described previously90, except using 1% SDS instead of 1% sarcosyl. The DNA embedded in the plugs was digested as previously described91, with adaptations. In brief, for each timepoint, one-third of an agarose plug was equilibrated four times in 5 ml of TE (10 mM Tris-HCl pH 7.5, 1 mM EDTA pH 7.5) for 15 min on a rotating wheel at room temperature. Agarose plugs were melted at 65 °C for 10 min, equilibrated at 42 °C for at least 20 min and digested with 0.5 U β-agarase I (New England Biolabs) at 42 °C for 45 min, followed by heat inactivation of β-agarase I at 65 °C for 15 min. After digestion with β-agarase I, the DNA was digested with 20 U of the respective restriction enzyme and an appropriate dilution of restriction enzyme buffer (rCutsmart, New England Biolabs) at 37 °C for 2 h. After 2 h, an additional 20 U of fresh restriction enzyme and half of the previously added volume of restriction enzyme buffer was added and the DNA was further digested at 37 °C for 2 h. Approximately 600 ng of digested DNA was loaded on a 0.8% agarose gel in 1× TBE and run at 2 V cm−1 for 14 h, using 30 ng of BstEII-digested lambda DNA (New England Biolabs) as a molecular mass standard. After DNA transfer to a GeneScreen Plus membrane (Revvity) by alkaline transfer, the membranes were hybridized with an [α−32P]dCTP-radiolabelled probe for the ERG1 or CCT6 hotspot and lambda DNA-BstEII digest (0.5 ng), exposed to a phosphor screen and imaged using the Amersham Typhoon phosphor imager (Cytiva). DSB signal intensities were quantified relative to the total lane signal using ImageQuant software. The signal at 0 h in SPM was used for background subtraction. The restriction enzymes used for digestion and the primer sequences used for probe amplification were as described previously91: ERG1, NcoI, (5′-CTGCCTACTCAAAACAGCAAAG, 5′-GTGAAGGAAGCACGTCAGAAAAAGC); CCT6, PstI, (5′-GCGTCCCGCAAGGACATTAG, 5′-TTGTGGCTAATGGTTTTGCGGTG).
Quantification and statistical analysis
All quantifications and data analyses were performed using Excel (Microsoft) and Prism (v.9.5.1 for macOS; GraphPad) software. The number of analysed cells (n) and the statistical analyses are described in each figure legend. For multiple comparisons, the non-parametric Kruskal–Wallis test was performed in Prism, followed by a post hoc Dunn’s test for multiple-comparison correction. For pairwise comparisons, two-tailed unpaired Mann–Whitney U-tests were used.
Ethics and inclusion statement
This study complies with the ethics and inclusion guidelines of Nature.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Relevant data supporting the findings of this study are provided within the Article and its Supplementary Information. Source data for all images, Southern blots and western blots are available at Zenodo92 (https://doi.org/10.5281/zenodo.15862742). Biological materials used in this study are available from the corresponding author on reasonable request.
References
Holliday, R. A mechanism for gene conversion in fungi. Genet. Res. 5, 282–304 (1964).
Schwacha, A. & Kleckner, N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83, 783–791 (1995).
Allers, T. & Lichten, M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57 (2001).
Zickler, D. & Kleckner, N. Meiosis: dances between homologs. Ann. Rev. Genet. 57, 1–63 (2023).
Ur, S. N. & Corbett, K. D. Architecture and dynamics of meiotic chromosomes. Ann. Rev. Genet. 55, 497–526 (2021).
Voelkel-Meiman, K., Moustafa, S. S., Lefrançois, P., Villeneuve, A. M. & MacQueen, A. J. Full-length synaptonemal complex grows continuously during meiotic prophase in budding yeast. PLoS Genet. 8, e1002993 (2012).
Rog, O., Köhler, S. & Dernburg, A. F. The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors. eLife 6, 4482 (2017).
Pattabiraman, D., Roelens, B., Woglar, A. & Villeneuve, A. M. Meiotic recombination modulates the structure and dynamics of the synaptonemal complex during C. elegans meiosis. PLoS Genet. 13, e1006670 (2017).
Nadarajan, S. et al. Polo-like kinase-dependent phosphorylation of the synaptonemal complex protein SYP-4 regulates double-strand break formation through a negative feedback loop. eLife 6, e23437 (2017).
Zhang, Z. et al. Multivalent weak interactions between assembly units drive synaptonemal complex formation. J. Cell Biol. 219, e201910086 (2020).
Padmore, R., Cao, L. & Kleckner, N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66, 1239–1256 (1991).
Henderson, K. A. & Keeney, S. Synaptonemal complex formation: where does it start? BioEssays 27, 995–998 (2005).
Hunter, N. & Kleckner, N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106, 59–70 (2001).
Börner, G. V., Kleckner, N. & Hunter, N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45 (2004).
Fung, J. C., Rockmill, B., Odell, M. & Roeder, G. S. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116, 795–802 (2004).
Chua, P. R. & Roeder, G. S. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93, 349–359 (1998).
Agarwal, S. & Roeder, G. S. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102, 245–255 (2000).
Novak, J. E., Ross-Macdonald, P. B. & Roeder, G. S. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158, 1013–1025 (2001).
Tsubouchi, T., Zhao, H. & Roeder, G. S. The meiosis-specific Zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with Zip2. Dev. Cell 10, 809–819 (2006).
Muyt, A. D. et al. A meiotic XPF-ERCC1-like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev. 32, 283–296 (2018).
Arora, K. & Corbett, K. D. The conserved XPF:ERCC1-like Zip2:Spo16 complex controls meiotic crossover formation through structure-specific DNA binding. Nucleic Acids Res. 47, 2365–2376 (2019).
Pyatnitskaya, A., Andreani, J., Guérois, R., Muyt, A. D. & Borde, V. The Zip4 protein directly couples meiotic crossover formation to synaptonemal complex assembly. Genes Dev. 36, 53–69 (2021).
Pyatnitskaya, A., Borde, V. & Muyt, A. D. Crossing and zipping: molecular duties of the ZMM proteins in meiosis. Chromosoma 128, 181–198 (2019).
Wojtasz, L. et al. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 5, e1000702 (2009).
Thacker, D., Mohibullah, N., Zhu, X. & Keeney, S. Homologue engagement controls meiotic DNA break number and distribution. Nature 510, 241–246 (2014).
Subramanian, V. V. et al. Chromosome synapsis alleviates Mek1-dependent suppression of meiotic DNA repair. PLoS Biol. 14, e1002369 (2016).
Voelkel-Meiman, K., Cheng, S.-Y., Morehouse, S. J. & MacQueen, A. J. Synaptonemal complex proteins of budding yeast define reciprocal roles in MutSγ-mediated crossover formation. Genetics 203, 1091–1103 (2016).
Mu, X., Murakami, H., Mohibullah, N. & Keeney, S. Chromosome-autonomous feedback down-regulates meiotic DNA break competence upon synaptonemal complex formation. Genes Dev. 34, 1605–1618 (2020).
Lee, M.-S. et al. The synaptonemal complex central region modulates crossover pathways and feedback control of meiotic double-strand break formation. Nucleic Acids Res. 49, 7537–7553 (2021).
Xu, L., Ajimura, M., Padmore, R., Klein, C. & Kleckner, N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15, 6572–6581 (1995).
Clyne, R. K. et al. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 5, 480–485 (2003).
Sourirajan, A. & Lichten, M. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 22, 2627–2632 (2008).
Matos, J., Blanco, M. G., Maslen, S., Skehel, J. M. & West, S. C. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147, 158–172 (2011).
Blanco, M. G., Matos, J. & West, S. C. Dual control of Yen1 nuclease activity and cellular localization by Cdk and Cdc14 prevents genome instability. Mol. Cell 54, 94–106 (2014).
Arter, M. et al. Regulated crossing-over requires inactivation of Yen1/GEN1 resolvase during meiotic prophase I. Dev. Cell 45, 785–800 (2018).
Sym, M. & Roeder, G. S. Zip1-induced changes in synaptonemal complex structure and polycomplex assembly. J. Cell Biol. 128, 455–466 (1995).
Humphryes, N. et al. The Ecm11-Gmc2 complex promotes synaptonemal complex formation through assembly of transverse filaments in budding yeast. PLoS Genet. 9, e1003194 (2013).
Voelkel-Meiman, K. et al. SUMO localizes to the central element of synaptonemal complex and is required for the full synapsis of meiotic chromosomes in budding yeast. PLoS Genet. 9, e1003837 (2013).
Yu, H.-G. & Koshland, D. Chromosome morphogenesis: condensin-dependent cohesin removal during meiosis. Cell 123, 397–407 (2005).
Prieler, S. et al. Spo11 generates gaps through concerted cuts at sites of topological stress. Nature 594, 577–582 (2021).
Jessop, L., Rockmill, B., Roeder, G. S. & Lichten, M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of Sgs1. PLoS Genet. 2, e155 (2006).
Wu, L. & Hickson, I. D. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870–874 (2003).
Muyt, A. D. et al. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46, 43–53 (2012).
Zakharyevich, K., Tang, S., Ma, Y. & Hunter, N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149, 334–347 (2012).
Leung, W.-K. et al. The synaptonemal complex is assembled by a polySUMOylation-driven feedback mechanism in yeast. J. Cell Biol. 211, 785–793 (2015).
Bermúdez-López, M. et al. Sgs1 roles in DNA end resection, HJ dissolution, and crossover suppression require a two-step SUMO regulation dependent on Smc5/6. Genes Dev. 30, 1339–1356 (2016).
Bonner, J. N. et al. Smc5/6 mediated sumoylation of the Sgs1-Top3-Rmi1 complex promotes removal of recombination intermediates. Cell Rep. 16, 368–378 (2016).
Texari, L. et al. The nuclear pore regulates GAL1 gene transcription by controlling the localization of the SUMO protease Ulp1. Mol. Cell 51, 807–818 (2013).
Carballo, J. A., Johnson, A. L., Sedgwick, S. G. & Cha, R. S. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132, 758–770 (2008).
Xu, L., Weiner, B. M. & Kleckner, N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 11, 106–118 (1997).
Hollingsworth, N. M. & Gaglione, R. The meiotic-specific Mek1 kinase in budding yeast regulates interhomolog recombination and coordinates meiotic progression with double-strand break repair. Curr. Genet. 65, 631–641 (2019).
Jordan, P. W., Karppinen, J. & Handel, M. A. Polo-like kinase is required for synaptonemal complex disassembly and phosphorylation in mouse spermatocytes. J. Cell Sci. 125, 5061–5072 (2012).
Argunhan, B. et al. Fundamental cell cycle kinases collaborate to ensure timely destruction of the synaptonemal complex during meiosis. EMBO J. 36, 2488–2509 (2017).
Tang, S. et al. Protecting double-Holliday junctions ensures crossing over during meiosis. Nature https://doi.org/10.1038/s41586-025-09555-1 (2025).
Cannavo, E. et al. Regulation of the MLH1-MLH3 endonuclease in meiosis. Nature 586, 618–622 (2020).
Kulkarni, D. S. et al. PCNA activates the MutLγ endonuclease to promote meiotic crossing over. Nature 586, 623–627 (2020).
Dernburg, A. F. et al. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94, 387–398 (1998).
McKim, K. S. et al. Meiotic synapsis in the absence of recombination. Science 279, 876–878 (1998).
Rosu, S. et al. The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet. 9, e1003674 (2013).
Stamper, E. L. et al. Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLoS Genet. 9, e1003679 (2013).
Machovina, T. S. et al. A surveillance system ensures crossover formation in C. elegans. Curr. Biol. 26, 2873–2884 (2016).
Matos, J. et al. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell 135, 662–678 (2008).
Benjamin, K. R., Zhang, C., Shokat, K. M. & Herskowitz, I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17, 1524–1539 (2003).
Haruki, H., Nishikawa, J. & Laemmli, U. K. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell 31, 925–932 (2008).
Yesbolatova, A. et al. The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nat. Commun. 11, 5701 (2020).
King, G. A. et al. Meiotic nuclear pore complex remodeling provides key insights into nuclear basket organization. J. Cell Biol. 222, e202204039 (2022).
Scherthan, H. et al. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 104, 16934–16939 (2007).
Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004).
Knop, M. et al. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963–972 (1999).
Morawska, M. & Ulrich, H. D. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast 30, 341–351 (2013).
Hentges, P., Van Driessche, B., Tafforeau, L., Vandenhaute, J. & Carr, A. M. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast 22, 1013–1019 (2005).
Oelschlaegel, T. et al. The yeast APC/C subunit Mnd2 prevents premature sister chromatid separation triggered by the meiosis-specific APC/C-Ama1. Cell 120, 773–788 (2005).
Grigaitis, R., Susperregui, A., Wild, P. & Matos, J. Characterization of DNA helicases and nucleases from meiotic extracts of S. cerevisiae. Methods Cell. Biol. 144, 371–388 (2018).
Wild, P. et al. Network rewiring of homologous recombination enzymes during mitotic proliferation and meiosis. Mol. Cell 75, 859–874 (2019).
Loidl, J., Klein, F. & Engebrecht, J. In Methods in Cell Biology, Vol. 53 (ed. Berrios M.) 257–285 (Academic, 1997).
Loidl, J., Nairz, K. & Klein, F. Meiotic chromosome synapsis in a haploid yeast. Chromosoma 100, 221–228 (1991).
Grigaitis, R. et al. Phosphorylation of the RecQ helicase Sgs1/BLM controls its DNA unwinding activity during meiosis and mitosis. Dev. Cell 53, 706–723 (2020).
Bommi, J. R. et al. Meiosis-specific cohesin component, Rec8, promotes the localization of Mps3 SUN domain protein on the nuclear envelope. Genes Cells 24, 94–106 (2019).
Shinohara, M., Oh, S. D., Hunter, N. & Shinohara, A. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 40, 299–309 (2008).
Voelkel-Meiman, K. et al. Crossover recombination and synapsis are linked by adjacent regions within the N terminus of the Zip1 synaptonemal complex protein. PLoS Genet. 15, e1008201 (2019).
Iwasaki, D. et al. The MRX complex ensures NHEJ fidelity through multiple pathways including Xrs2-FHA–dependent Tel1 activation. PLoS Genet. 12, e1005942 (2016).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
ImageJ plugin HyperStackReg v.5.6 (Zenodo, 2018).
Salah, S.-M. & Nasmyth, K. Destruction of the securin Pds1p occurs at the onset of anaphase during both meiotic divisions in yeast. Chromosoma 109, 27–34 (2000).
Martini, E., Diaz, R. L., Hunter, N. & Keeney, S. Crossover homeostasis in yeast meiosis. Cell 126, 285–295 (2006).
Kim, K. P. et al. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143, 924–937 (2010).
Ahuja, J. S. & Borner, G. V. Analysis of meiotic recombination intermediates by two-dimensional gel electrophoresis. Methods Mol. Biol. 745, 99–116 (2011).
Owens, S., Tang, S. & Hunter, N. Monitoring recombination during meiosis in budding yeast. Methods Enzymol. 601, 275–307 (2018).
Schwacha, A. & Kleckner, N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90, 1123–1135 (1997).
Murakami, H., Borde, V., Nicolas, A. & Keeney, S. Gel electrophoresis assays for analyzing DNA double-strand breaks in Saccharomyces cerevisiae at various spatial resolutions. Methods Mol. Biol. 557, 117–142 (2009).
Pan, J. et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144, 719–731 (2011).
Henggeler, A., Orlić, L., Velikov, D. & Matos, J. Data for ‘Holliday junction–ZMM protein feedback enables meiotic crossover assurance’. Zenodo https://doi.org/10.5281/zenodo.15862742 (2025).
Acknowledgements
We thank A. Shinohara, A. MacQueen, A. Pichler, K. Weis, M. Shinohara, F. Stutz and F. Klein for reagents; M. Arter for initial work characterizing Yen1ON; N. Hunter for communicating unpublished data; V. Borde and M. Jagannathan for advice; V. Jantsch, M. Blanco and M. Xaver for reading the manuscript; and the staff at the Max Perutz Labs BioOptics facility for providing support for microscopy experiments. The Matos Lab was supported by the Swiss National Science Foundation SNSF (176108), the Austrian Science Foundation FWF (SFB Meiosis, 8807-B; 10.55776/F88) and the European Research Council ERC (101002629).
Funding
Open access funding provided by University of Vienna.
Author information
Authors and Affiliations
Contributions
A.H. performed all experiments except for the Southern blots performed in Figs. 1–3 and Extended Data Figs. 3, 5h and 6, which were performed by L.O., and the live-cell imaging experiments in Fig. 3 and Extended Data Figs. 1 and 7, which were performed by D.V.; J.M. conceived and supervised the project. A.H. and J.M. wrote the manuscript with input from all of the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 HJs stabilize the SC during meiotic pachytene.
a, Representative fluorescence-activated cell sorting (FACS) analysis of DNA content at regular time intervals after induction of meiosis in SPM to monitor pre-meiotic DNA replication in cells with the indicated genotypes. Note that most cells have replicated their genomes after ~4 h in SPM. Corresponds to experiments in Fig. 1b–g and Extended Data Fig. 1b–g. b, c, Western blot analysis (b) and quantification of Yen1ON and Zip1 protein levels normalized using Pgk1 as loading control (c) in cells at indicated times in SPM. Yen1ON was induced by β-oestradiol addition (or MeOH as control) at 7 h. Corresponds to the experiment in Fig. 1b,c and Extended Data Fig. 1f. Yen1ON protein levels after addition of β-oestradiol are also shown in Fig. 1d. d, e, As in (b) and (c), for PGAL1-Yen1ON-ND. Corresponds to the experiment in Fig. 1e,f and Extended Data Fig. 1g. Yen1ON-ND protein levels after addition of β-oestradiol are also shown in Fig. 1g. f, Representative images of meiotic chromosome spreads at indicated times in SPM (left) and quantification of Zip1 synapsis and polycomplex formation (right) in the absence of Yen1ON induction (MeOH-treated control) at 7 h (n = 50 nuclei per time point; representative of three biological replicates). Control for the experiment shown in Fig. 1b–d. g, As in (f), for PGAL1-Yen1ON-ND. Control for the experiment shown in Fig. 1e–g. h, i, Time-lapse image montage of Zip1GFP in a cell nucleus (h) and quantification of structured Zip1GFP signal (i) after Yen1ON induction by β-oestradiol addition at ~7 h in SPM (t = 0 min). Arrowhead marks the emerging polycomplex. Mean of two biological replicates is plotted (n = 40 cells each). Corresponds to Supplementary Video 1. Scale bar = 1 µm. j, k, As in (h) and (i), for PGAL1-Yen1ON-ND. Mean of two biological replicates is plotted (n = 40 cells each). Corresponds to Supplementary Video 2.
Extended Data Fig. 2 HJs stabilize the SC central region but are dispensable for the axis-loop organization of pachytene chromosomes.
a, Representative images of meiotic chromosome spreads at indicated times in SPM, immunostained for Ecm11-Gmc2 (green) and Smt3 (blue). Yen1ON expression was induced by β-oestradiol addition (or MeOH as control) at 7 h in SPM. Scale bars = 2 µm. b, c, Quantification of Ecm11-Gmc2 synapsis (b) and SC-associated Smt3 (c) from (a) (n = 50 nuclei per time point; representative of two biological replicates). d, Representative images of meiotic chromosome spreads at indicated times in SPM, immunostained for Zip1 (green) and Rec8 (magenta). Yen1ON expression was induced by β-oestradiol addition (or MeOH as control) at 7 h in SPM. Scale bars = 2 µm. e, Quantification of chromosome axis morphology based on Rec8 immunostaining from (d) (n = 50 nuclei per time point; representative of two biological replicates). f, Representative STED microscopy images of meiotic chromosome spreads at indicated times in SPM, immunostained for Zip1 (green) and Rec8 (magenta). Yen1ON expression was induced by β-oestradiol addition (or MeOH as control) at 7 h in SPM. Insets show magnified regions indicated by dashed lines. Arrowheads indicate example chromosome regions with split axes. Scale bars = 1 µm; scale bars in insets = 200 nm.
Extended Data Fig. 3 Recombination intermediates containing dHJs and SEIs are stable during pachytene.
a, Representative in situ immunofluorescence images of cells at indicated times in SPM after Rec104FRB anchor-away by rapamycin addition (or DMSO as control) at 7 h, immunostained for the HA epitope to visualize the subcellular localization of Rec104FRB. DNA is visualized with DAPI (grey). b, Quantification of cells with nuclear Rec104FRB at indicated times in SPM from (a) (n = 100 cells per time point). c, Southern blot analysis of recombination at the HIS4::LEU2 recombination hotspot from the experiment described in Fig. 1h. Two biological replicates are shown; replicate 1 corresponds to the cropped blot in Fig. 1i. P1, parental 1; P2, parental 2; CO1, crossover 1; CO2, crossover 2; DSBs, double-strand breaks; JMs, DNA joint molecules. Asterisk indicates meiosis-specific recombinant band from gene conversion of the XhoI site closest to the DSB site. Dagger indicates ectopic recombination bands between HIS4::LEU2 and leu2::hisG at the native LEU2 locus. d, Native/native two-dimensional Southern blot analysis of branched JMs at the HIS4::LEU2 recombination hotspot from indicated time points of replicate 1 in (c). The different DNA JM species are depicted in the left blot image. SEI, single-end intermediate; IH-dHJ, interhomologue-dHJ; IS-dHJ, intersister-dHJ; mcJM, multichromatid DNA JM. e, Quantification of SEI, IH-dHJ, IS-dHJ and mcJM levels from (d), shown as the percentage of total DNA. f, Western blot analysis of Yen1ON protein levels in cells from indicated times in SPM of replicate 1 in (c). Crm1 served as protein loading control. Representative of two biological replicates. g, Quantification of Yen1ON protein levels from the Western blot in (f) and a biological replicate, normalized to the highest value (red line), along with mean JM levels from both replicates in (c) (grey line). Error bars represent the range.
Extended Data Fig. 4 ZMMs are continuously required to maintain chromosome synapsis during prophase I.
a, Representative images of meiotic chromosome spreads at indicated times in SPM following Yen1ON induction by β-oestradiol addition (or MeOH as control) at 7 h, immunostained for Ecm11-Gmc2 (green) and Msh5 (magenta). Scale bars = 2 µm. b, Quantification of Msh5 foci number from (a) (mean ± s.d.; n = 30 nuclei per time point; Kruskal-Wallis test (p ≤ 0.001) with Dunn’s multiple comparison test). Representative of two biological replicates. c, d, As in (a) and (b), for analysis of Zip49myc (mean ± s.d.; n = 30 nuclei per time point; Kruskal-Wallis test (p ≤ 0.001) with Dunn’s multiple comparison test). Representative of two biological replicates. e, Quantification of Zip3, Msh5 and Zip49myc aggregates associating with Ecm11-Gmc2 polycomplexes at indicated times in SPM. Yen1ON was induced by β-oestradiol addition (or MeOH as control) at 7 h (n = 30 nuclei per time point; representative of two biological replicates). f, g, Western blot analysis of Msh4AID (f) and Zip4AID (g) protein levels in cells at indicated times in SPM. OsTir1F74G expression was induced by addition of CuSO4 at 6.5 h, and AID-dependent depletion was induced by 5-Ph-IAA addition (or DMSO as a control) at 7 h, as described in Fig. 2c. Crm1 served as protein loading control. Representative of two biological replicates. h, Representative images of meiotic chromosome spreads at indicated times in SPM from (f), immunostained for Zip1 (green) and Msh4AID (magenta). Scale bars = 2 µm. i, As in (h), for ZIP4AID in (g). j, k, Quantification of Zip1 synapsis and polycomplexes from (h) and (i) (n = 50 nuclei per time point; representative of two biological replicates). l, Representative images of meiotic chromosome spreads at indicated times in SPM, immunostained for Zip1 (green) and Rec8 (magenta). Zip3AID depletion was induced by 5-Ph-IAA addition (or DMSO as control) at 7 h. Scale bars = 2 µm. m-o, Quantification of chromosome axis morphology based on Rec8 immunostaining in cells with indicated genotypes at specified times in SPM. Zip3AID, Msh4AID or Zip4AID depletion was induced by 5-Ph-IAA addition (or DMSO as control) at 7 h (n = 50 nuclei per time point; representative of two biological replicates).
Extended Data Fig. 5 ZMMs protect dHJs from Sgs1-mediated dissolution throughout prophase I.
a, Western blot analysis of Msh4AID protein levels in cells at indicated times in SPM. OsTir1F74G expression was induced by addition of CuSO4 at 6.5 h, and Msh4AID depletion was induced by 5-Ph-IAA addition (or DMSO as a control) at 7 h, as described in Fig. 2c. Crm1 served as protein loading control. Representative of two biological replicates. b, c, Southern blot (b) and quantification of JMs, non-crossover (NCO1) and crossover (CO2) products (c) at the HIS4::LEU2 recombination hotspot from (a). d, As in (a), for MSH4AID SGS1AID. Representative of two biological replicates. e,f, As in (b) and (c), for MSH4AID SGS1AID from (d). g, As in (a), for SGS1AID. h, i, As in (b) and (c), for SGS1AID from (g). j, Representative images of meiotic chromosome spreads at indicated times in SPM from (d), immunostained for Zip1 (green) and Msh4AID/Sgs1AID (magenta). Scale bars = 2 µm. k, Quantification of Zip1 synapsis and polycomplexes from (j) (n = 50 nuclei per time point; representative of two biological replicates). l, Time-lapse image montage of Zip1GFP in cell nuclei of indicated genotypes. OsTir1F74G expression was induced by addition of CuSO4 at 6.5 h in SPM, and Zip3AID and Sgs1AID depletion was induced by 5-Ph-IAA addition (or DMSO as a control) at ~7 h in SPM (t = 0 min). Arrowhead marks the emerging polycomplex. Corresponds to Supplementary Video 3 and 4. Scale bar = 1 µm. m, Quantification of structured Zip1GFP signal in cells of indicated genotypes from (l) (n = 40 cells per genotype). n, Quantification of time to complete loss of structured Zip1GFP signal in cells of indicated genotypes from (m) (mean ± s.d; n = 40 cells per genotype; two-tailed, unpaired Mann-Whitney U test). o, As in (a), for ZIP4AID SGS1AID. Representative of two biological replicates. p, As in (j), for (o). q, As in (k), for (p) (n = 50 nuclei per time point; representative of two biological replicates).
Extended Data Fig. 6 A molecular tool to reversibly induce SC disassembly independent of HJ resolution.
a, Ulp1 domain organization, adapted from48. In the ulp1∆N mutant, an in-frame GFP coding sequence replaces parts of the nuclear pore complex anchoring region (amino acids 172–340) in the N-terminal domain. b, Schematic representation of the ULP1 genomic locus. A diploid yeast strain heterozygous for the ulp1∆N-FRB allele was used since ULP1 is an essential gene. The native promoter of ulp1∆N-FRB is replaced by PGAL1, which can be induced by the addition of β-oestradiol. In addition, the C-terminal FRB domain of ulp1∆N-FRB enables the conditional nuclear depletion using the rapamycin-dependent anchor-away system. c, Western blot analysis of Ulp1∆N-FRB, Smt3, Ecm11 and Zip1 protein levels in cells at indicated times in SPM. Ulp1∆N-FRB expression was induced by β-oestradiol addition (or MeOH as control) at 8 h, and nuclear depletion was induced by rapamycin addition (or DMSO as control) at 9 h. Arrow indicates the band corresponding to the expected molecular weight of Ulp1∆N-FRB. Asterisks indicate putative SUMOylated and polySUMOylated Ecm11 bands in the anti-Smt3 blot. Samples were run on separate gels due to similar molecular masses of the proteins, with Pgk1 as a sample processing control. Representative of two biological replicates. d, Quantification of SC-associated Smt3 on meiotic chromosome spreads from (c) (n = 50 nuclei per time point; representative of two biological replicates). e, Southern blot analysis of JMs and DSBs at the HIS4::LEU2 recombination hotspot. Ulp1∆N was induced by β-oestradiol addition (or MeOH as control) at 7 h in SPM. Two biological replicates are shown, with replicate 2 including an additional time point (12 h in SPM). Replicate 1 corresponds to the cropped blot in Fig. 3d. f, Representative STED microscopy images of meiotic chromosome spreads at indicated times in SPM from (c), immunostained for Zip1 (green) and Rec8 (magenta). Insets show magnified regions indicated by dashed lines. Note that although Zip1 is lost from chromosomes and Rec8 localization appears to be more discontinuous after Ulp1∆N expression (see also (g)), the co-alignment of homologous axes is maintained. Scale bars = 1 µm; scale bars in insets = 200 nm. g, Quantification of chromosome axis morphology based on Rec8 immunostaining on meiotic chromosome spreads at indicated times in SPM from (c) (n = 50 nuclei per time point; representative of two biological replicates). h, Experimental setup to determine whether SC reassembly upon Ulp1∆N-FRB expression/nuclear depletion depends on the formation and repair of de novo DSBs. i, Western blot analysis of Ulp1∆N-FRB, Rec104AID, Smt3, Ecm11 and Zip1 protein levels in cells at indicated times in SPM, treated as described in (h). Arrow indicates the band corresponding to the expected molecular weight of Ulp1∆N-FRB. Asterisks indicate putative SUMOylated and polySUMOylated Ecm11 bands in the anti-Smt3 blot. Samples were run on separate gels due to similar molecular masses of the proteins, with Pgk1 as a sample processing control. j, Quantification of Zip1 synapsis on meiotic chromosome spreads from (i) (n = 50 nuclei per time point).
Extended Data Fig. 7 dHJ-ZMM protein interplay enables SC reassembly upon Ulp1∆N-FRB depletion.
a, Western blot analysis of Ulp1∆N, Yen1ON and Ecm11 protein levels in cells with the indicated genotype at specified times in SPM, treated as described in Fig. 3f. Arrow indicates the band corresponding to the expected molecular weight of Ulp1∆N-FRB. Samples were run on separate gels due to similar molecular masses of the proteins, with Pgk1 as a sample processing control. b, Representative images of meiotic chromosome spreads of the indicated genotype from (a) at specified times in SPM, immunostained for Zip1 (green). Control for the experiment shown in Fig. 3g. Scale bars = 2 µm. c, Quantification of Zip1 synapsis from (b) (n = 50 nuclei per time point). d, Time-lapse image montage of Zip1GFP in a cell nucleus after Ulp1∆N-FRB expression induced by β-oestradiol addition at 7 h in SPM (t = 0 min). Control for the experiment shown in Fig. 3i, with DMSO added after 120 min. Corresponds to Supplementary Video 5. Scale bar = 1 µm. e, Quantification of structured Zip1GFP signal from (d). Mean of two biological replicates is plotted (n = 40 cells each). f, Representative images of meiotic chromosome spreads at indicated times in SPM, immunostained for Zip3 (green) and Smt3 (magenta). Ulp1∆N-FRB expression was induced by β-oestradiol addition at 7 h in SPM, and nuclear depletion was initiated by rapamycin addition at 9 h in SPM. Arrowheads mark Zip3 aggregates. Scale bars = 2 µm. g, h, Quantification of Zip3 foci number (g) and fraction of nuclei exhibiting Zip3 aggregates (h) from (f) (mean ± s.d.; n = 30 nuclei per time point; Kruskal-Wallis test (p ≤ 0.001) with Dunn’s multiple comparison test). i, As in (f), for the simultaneous expression of Ulp1∆N-FRB and Yen1ON by β-oestradiol addition at 7 h in SPM. j, k, As in (g) and (h), for (i) (mean ± s.d.; n = 30 nuclei per time point; Kruskal-Wallis test (p ≤ 0.001) with Dunn’s multiple comparison test).
Extended Data Fig. 8 dHJ-ZMM protein interplay suppresses DSB formation.
a, Experimental setup to measure DSB formation by Southern blotting after Yen1ON expression in ndt80∆ cells. Sae2AID is depleted by 5-Ph-IAA addition (or DMSO as control) alongside Yen1ON induction by β-oestradiol addition (or MeOH as control) to prevent endonucleolytic removal of Spo11 and subsequent DSB processing and repair. b, Western blot analysis of Yen1ON, Sae2AID and Hop1 protein levels in cells at indicated times in SPM, treated as described in (a). Hop1P indicates phosphorylated Hop1. Crm1 served as protein loading control. Representative of two biological replicates. c, d, Southern blot analysis of DSB formation at the CCT6 (c) and ERG1 (d) hotspots from (b). Two biological replicates are shown side-by-side in the same blot for each hotspot; replicate 2 of the CCT6 blot corresponds to the cropped blot in Fig. 4d. e, f, Southern blot analysis of DSB formation at the CCT6 (e) and ERG1 (f) hotspots in cells with the indicated genotype, with Msh4AID depletion by 5-Ph-IAA addition (or DMSO as control) at 7 h in SPM. Two biological replicates are shown per locus; replicate 1 of the CCT6 blot in (e) corresponds to the cropped blot in Fig. 4g. g, Western blot analysis of Zip3AID, Hop1 and Hop1-pT318 protein levels in cells at indicated times in SPM. Zip3AID depletion was induced by 5-Ph-IAA addition (or DMSO as control) at 7 h in SPM. Hop1P indicates phosphorylated Hop1. Pgk1 served as protein loading control. h, As in (g), for ZIP4AID.
Extended Data Fig. 9 Premature resolution of recombination intermediates disrupts the SC and causes meiosis I failure.
a, Representative FACS analysis of DNA content in cells with the indicated genotype at regular time intervals after induction of meiosis in SPM. Corresponds to the experiment in Fig. 4i–l and Extended Data Fig. 9b–l. b, Time-lapse image montage of Zip1GFP in a cell nucleus induced to enter meiosis in SPM, with imaging starting at 4.5 h in SPM (t = −30 min). The durations of the zygotene and pachytene stages of prophase I, and the time from pachytene exit to anaphase I are indicated (mean ± s.d of two biological replicates; n = 20 cells each). The brightness of Nup84mCherry was adjusted to facilitate visibility of the signal. Asterisk indicates the gametogenesis uninherited nuclear compartment (GUNC). Ana, Anaphase. Corresponds to Supplementary Video 8. Scale bar = 1 µm. c, d, Western blot analysis (c) and quantification (d) of Yen1ON protein levels, normalized to the highest value, in cells at indicated times after Yen1ON induction by β-oestradiol addition (or MeOH as control) at 7 h in SPM. Error bars represent range of two biological replicates. e, Time-lapse image montage of Zip1GFP in a cell nucleus after MeOH addition as a control in early to mid-zygotene (t = 0 min, ~5 h in SPM). The brightness of Nup84mCherry was adjusted to facilitate visibility of the signal. Corresponds to Supplementary Video 10. Scale bar = 1 µm. f, Quantification of structured Zip1GFP signal from (e). Mean of two biological replicates is plotted (n = 20 cells each). g-i, Quantification of structured Zip1GFP signal (g), meiosis I entry (h), and sporulation (i) in pachytene cells at the time of β-oestradiol addition (t = 0 min, ~5 h in SPM). Mean of two biological replicates is plotted (n = 20 cells each; error bars represent range). Corresponds to Supplementary Video 11. j-l, Quantification of structured Zip1GFP signal (j), meiosis I entry (k), and sporulation (l) in pre-leptotene/leptotene cells at the time of β-oestradiol addition (t = 0 min, ~5 h in SPM). Mean of two biological replicates is plotted (n = 20 cells each; error bars represent range). Corresponds to Supplementary Video 12. m, Time-lapse image montage of Zip1GFP and Htb1mCherry in a cell nucleus of the indicated genotype. H2O was added as a control at 7 h in SPM (t = −65 min). Ndt80 expression was induced by β-oestradiol addition at ~8 h in SPM (t = 0 min). Corresponds to Supplementary Video 13. Scale bar = 1 µm. n, Quantification of structured Zip1GFP signal and the fraction of cells undergoing meiosis I and II based on Htb1mCherry labelled chromatin from (m) (n = 60 cells). o, As in (m), with CuSO4 addition to induce expression of Yen1ON at 7 h in SPM (t = −65 min). Corresponds to Supplementary Video 14. p, As in (n), for (o). q, Quantification of cells with completed or defective meiotic divisions from (n) and (p).
Extended Data Fig. 10 Cdc5 promotes SC disassembly in part through the activation of HJ resolvases.
a, Quantification of Zip1 synapsis and polycomplexes in the absence of Cdc5 induction (MeOH-treated control) at 7 h in SPM (n = 50 nuclei per time point). Controls for the experiments shown in Fig. 5a,b. b, Western blot analysis of Cdc5 and Zip1 protein levels in cells of indicated genotypes at specified times in SPM. Cdc5 was induced by β-oestradiol addition (or MeOH as control) at 7 h in SPM. Pgk1 served as protein loading control. Correspond to experiments in (a) and Fig. 5a,b. c, Quantification of Zip1 protein levels from (b), normalized to the highest value. d, Representative images of meiotic chromosome spreads at indicated times in SPM, immunostained for Zip1 (green). Cdc5 was induced by β-oestradiol addition (or MeOH as control) at 7 h in SPM. Scale bars = 2 µm. e, Quantification of Zip1 synapsis and polycomplexes from (d) (n = 50 nuclei per time point). f, As in (b), for cells of the indicated genotype. Corresponds to experiment in (d) and (e). g, As in (c), for (f). h, i, Time-lapse image montage of Zip1GFP in cell nuclei of indicated genotypes (h) and quantification of structured Zip1GFP signal (i) after Cdc5 induction by β-oestradiol addition at ~7 h in SPM (t = 0 min) (n = 50 cells per genotype). The bottom panel corresponds to Supplementary Video 17. Scale bars = 1 µm. j, Quantification of time to complete loss of structured Zip1GFP signal in cells of indicated genotypes from (i) (mean ± s.d; n = 50 cells per genotype; two-tailed, unpaired Mann-Whitney U test). k, (Top) Representative images of meiotic chromosome spreads of the indicated genotypes at 6 h in SPM, immunostained for Zip1 (green), Rec8 (magenta) and the SPB component γ-tubulin/Tub4 (cyan). Insets show magnified, slightly separated SPBs. Scale bars = 2 µm. (Bottom) Quantification of nuclei with partial or full SPB separation containing Zip1 stretches (weighted mean ± weighted SD of two biological replicates; n = 152–174 nuclei per condition).
Supplementary information
Supplementary Fig. 1
The uncropped Southern and western blot images generated in this study.
Supplementary Tables
A list of yeast strains used in this study (Supplementary Table 1) and the spore viability data for newly generated yeast strains to evaluate the functional impact of protein tagging (Supplementary Table 2).
Supplementary Information
The legends to Supplementary Videos 1–17.
Supplementary Videos
Supplementary Videos 1–17.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Henggeler, A., Orlić, L., Velikov, D. et al. Holliday junction–ZMM protein feedback enables meiotic crossover assurance. Nature (2025). https://doi.org/10.1038/s41586-025-09559-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41586-025-09559-x