Abstract
The diversity and biochemical potential of the PiggyBac transposase gene insertion system remains largely unexplored. Using a eukaryotic transposon mining pipeline, we expand the explored diversity by two orders of magnitude and experimentally validate a subset of highly divergent PiggyBac sequences. Fine-tuning a protein language model to further expand PiggyBac sequence space discovers transposases with improved activity and that are compatible with T cell engineering and Cas9-directed transposase-assisted integration.
Similar content being viewed by others
Main
The advancement of genome-engineering technologies has transformed biological engineering and opened new avenues for therapeutic and biotechnological applications1. Central to these developments are tools that enable efficient insertion of large DNA sequences into target genomes, an essential capability to unlock the full potential of synthetic biology2,3. Among these tools, DNA transposons have been widely adapted for genome modification across numerous organisms4,5. Notably, the PiggyBac transposase has emerged as a powerful tool because of its ability to integrate substantial DNA cargo across diverse cellular environments, making it a highly versatile platform for gene insertion.
Active PiggyBac elements have been identified in the genomes of insects and bats6,7 and phylogenetic studies have identified PiggyBac transposases across multiple eukaryotic families8,9. Nonetheless, much of their evolutionary diversity and biochemical potential remain unexplored. Traditionally, exploring PiggyBac diversity can be achieved by bioprospecting natural sequences. However, recent advances in generative artificial intelligence (AI) methods applied to protein design have shown that sampled natural diversity can be augmented to generate functional sequences not seen in nature10,11,12. For instance, a combination of RFdiffusion13 and methodologies to design catalytic sites created active synthetic serine hydrolases with new folds14. A protein large language model (pLLM) was recently used to generate a CRISPR–Cas9 that does not exist in nature but performs well for gene-editing applications10. The development of such models has opened up exciting opportunities to expand biodiversity and improve gene integration tools. Despite this broad exploration, the potential of PiggyBac as a gene insertion tool remains constrained by its preference for TTAA integration sites, limiting its target specificity and precision15. Efforts to improve targeting precision have explored fusions with engineered DNA-binding domains such as transcription activator-like effector, engineered zinc-finger proteins and CRISPR catalytically inactive Cas9, each with varying targeting efficiencies16,17,18. Our phylogenetic mining uncovered over 13,000 PiggyBac elements, revealing domain acquisitions across multiple PiggyBac clusters. We experimentally validated a subset of these elements, identifying ten active transposases with up to 30% sequence identity to one another, thereby expanding the functional repertoire of known PiggyBac elements. Additionally, we generated ‘mega-active’ synthetic variants of the widely used laboratory-evolved hyperactive PiggyBac (HyPB) transposase using a fine-tuned pLLM, Progen2 (ref. 19), and demonstrated the applicability of these PiggyBac orthologs in critical gene-editing contexts, such as primary T cell engineering and Cas9-directed transposase-assisted integration.
We searched all available eukaryotic genome assemblies on the National Center for Biotechnology Information (NCBI; 31,565 genomes) and Dfam20 (20,638 PiggyBac sequences) databases, finding a total of 273,643 PiggyBac transposon open reading frames (ORFs) together with their DNA sequences (Fig. 1a and Supplementary Fig. 1). To differentiate active transposons from transposase-derived proteins co-opted by the host that have lost transposition activity21,22, we retrieved sequences with the presence of an RNase H-like domain, cysteine-rich domain (CRD), terminal inverted repeats (TIR) and a target site duplication (TSD) with the TTAA motif (Supplementary Fig. 1). These motifs are reported to be crucial for DNA excision and integration7. Filtering yielded a dataset of 116,216 putatively transposition competent PiggyBac elements that resulted in 13,693 PiggyBac subfamilies after clustering at 80% sequence identity.
a, PiggyBac identification and testing pipeline overview (detailed pipeline in Supplementary Fig. 1 and Methods). Piggybac domains: N terminus, gray; double DNA-binding domain, yellow; catalytic domain, green; insertion domain, blue; CRD, pink (detailed domain depiction in Supplementary Fig. 7). Panel a created with BioRender. b, PiggyBac phylogenetic tree from the 2,500 identified clusters at 0.6 identity. Cluster size is represented by the circle radius on top of tree leaves and the number of unique taxonomic species present in the cluster is shown by circle color. Tree ring labels, from inner to outer: (1) identified PiggyBac main groups (five in total); (2) major cluster taxonomic groups; (3) clusters with more than one broad taxonomic group; (4) CRD classification; and (5) clusters with fusion domains. Tested PiggyBac clusters are marked with arrows, inactive PiggyBac clusters are marked with orange arrows and active PiggyBac clusters are marked with green arrows. The four colored stars represent previously described PiggyBac-like transposons with demonstrated autonomous activity: PiggyBat6, blue; PiggyBac5, red; Mage32, orange; PLE-wu33, purple. The ‘fish’ category includes Chondrichthyes, Agnatha and Osteichthyes (complete legends and colors in Supplementary Fig. 2). c, Experimental validation of PiggyBac orthologs by nontargeted transposon integration fluorescence assay in HEK293T cells 2 weeks after transfection, in the presence (TPS, pink) or absence (no TPS, green) of transposase plasmid. Data are presented as the mean values ± 95% confidence interval (CI), with n = 2 for orthologs with a mean level of RFP lower than 1% and n = 3 for those with higher (seven top performers). d, Sequence identity heat map between active orthologs from c. e, Effect of N-terminal phosphorylation substitutions on excision, measured by transposon excision fluorescence assay. StA indicates serine-to-alanine substitutions in CKII phosphorylation sites (Supplementary Fig. 7). Data are presented as the mean values relative to WT ± 95% CI, with n = 3. f, Targeted transposon integration qPCR assay with Poetur and Antgra4 orthologs in the triple-mutant background (R372A;K575A;D450N) at the AAVS1-3 site. Data are presented as the mean values ± 95% CI, with n = 1. g, Pictures of species containing the top two PiggyBac hits34. Credits: A. grandis, photo courtesy of USDA Agricultural Research Service; P. turrubarensis, Paradise Costa Rica.
The eukaryotic distribution of PiggyBac transposons is notably diverse, encompassing taxa from fungi and plants to mammals (Fig. 1b and Supplementary Fig. 2b); it is predominantly represented in insects (~60%), followed by fish and mollusks (5%). We identified five main PiggyBac groups (Fig. 1b and Supplementary Figs. 2a and 3a) on the basis of main tree phylogenetic branches, taxonomic distribution and the CRD types. More than 200 clusters are represented by more than one broad taxonomic group (Fig. 1b, ring 3), indicating widespread horizontal gene transfer across groups, as previously reported in other transposable elements23. Group 4 has a unique, unexpected taxonomic distribution with presence in fungi, land plants and algae (Fig. 1b, ring 1, purple). We also observed ‘superhost’ species, characterized by containing numerous PiggyBac sequences. The top three superhosts captured 7.3% of all PiggyBac diversity (Supplementary Fig. 3). Additionally, we found multiple domain acquisition events at both N and C termini, with 4.6% of all the reported clusters containing a fusion domain and N-terminal fusions being more predominant (Fig. 1b, ring 5). DNA-binding domains and fusogens were the most abundantly acquired domains, suggesting multiple transposition mechanisms for DNA recognition and cell entry (Supplementary Fig. 4).
We used AlphaFold3 (ref. 24) structural prediction and clustering to further understand the diversity of the CRD domain. We identified two main CRD cross brace zinc-finger folds, HC6H and C5HC2 (Supplementary Fig. 5). In contrast to C5HC2, the HC6H group is longer and retains two unique β-sheets in its insertion domain. The insertion domain consists of structures with three and five β-strands in C5HC2 and HC6H, respectively, which interrupts the catalytic domain after the seventh β-strand. While the catalytic domain catalyzes the hydrolysis and transesterification steps necessary for transposition, the insertion domain has a role in DNA binding and transposon integration7. Analysis of the catalytic domain indicates high structural conservation (root-mean-square deviation (r.m.s.d.) of the catalytic region near 2 Å and a template modeling (TM)-score of 0.915) despite high sequence divergence (Supplementary Fig. 5).
To explore the potential of bioprospected transposon diversity for gene insertion, we selected 23 representative PiggyBac sequences across the phylogenetic tree for experimental testing (Fig. 1b, colored triangles). These sequences were chosen to encompass all five major PiggyBac groups, both primary CRD types and a representative range of taxonomic groups. Transposition activity was validated through detecting excision of the transposase plasmid (Supplementary Fig. 6a) and nontargeted integration of a red fluorescent protein (RFP)-containing transposon payload in HEK293T cells (Fig. 1c). Nontargeted integration refers to the canonical PiggyBac transposition mechanism, in which it excises and inserts itself into TTAA motifs throughout the genome25. Of the tested sequences, nine (~40%) had detectable activity, with two sequences equivalent to laboratory-evolved HyPB5. Active sequences were spread across phylogeny and had low sequence identity to HyPB (Fig. 1d). This broad distribution of active elements across taxonomic and CRD diversity underscores the potential of PiggyBac transposons as versatile tools in genetic engineering and gene-transfer applications. Interestingly, the previously described PiggyBat sequence did not exhibit activity, which contrasts with previous reports6. This discrepancy is likely because of the fact that a consensus PiggyBat sequence generated in this study is constructed from multiple PiggyBat cluster sequences and is different from the previously described. To further improve transposon activity, we identified and removed CKII phosphorylation motifs in the N terminus of PiggyBac, previously reported to inhibit its transposition activity in HyPB7 (Supplementary Fig. 7b). CKII site removal increased transposition activity in both orthologs (Fig. 1e). We also tested how TIR truncation affected excision in Poetur and Antgra4 (Supplementary Fig. 8), identifying minimal TIR versions with equal activity. We further tested compatibility of our orthologs with the previously described FiCAT18 targeted insertion system. In the FiCAT platform, a Cas9 enzyme fused to an engineered Piggybac transposase induces a double-strand break (DSB) at a target genomic site. The PiggyBac component, engineered to be excision competent and integration deficient, excises a transposon delivered by plasmid. This transposon is then inserted into the DSB site, generating an integration signature mediated by nonhomologous end joining. Our results showed successful FiCAT compatibility of Poetur and Antgra4 in HEK293T cells (Fig. 1f and Supplementary Figs. 9 and 10).
Next, we sought to explore how the generated corpus of natural sequences could be used to improve the activity of existing transposases. We fine-tuned the ProGen2-base language model19 using over 13,000 bioprospected sequences, similarly to the method previously described for Cas9 nucleases10. In our training data, the HyPB sequence was included five of ten times, depending on the model, to bias the model toward improvement of the HyPB sequence. We created two separate models: one model to generate sequences from the N terminus to C terminus and the second to generate sequences from the C terminus to N terminus. We then generated over 100,000 sequences from these two models prompted with the first 50 (N–>C) or last 50 (C–>N) amino acids. A total of 50 amino acids were selected to give sufficient context to the models so that they could generate similar sequences, without giving so much that the model could perfectly recreate the HyPB sequence. Sequences were first filtered on the basis of a set of basic protein properties in addition to PiggyBac-specific properties (Fig. 2a and Supplementary Fig. 8b). We further filtered and scored sequences by structural (predicted local distance difference test (pLDDT), r.m.s.d. to experimental structure, SURFMAP26,27 and TM-scores) and deep learning scores (Progen perplexity, ProteinMPNN28 and ESM1v29). Generated sequences had higher pLDDT, ESM1v and ProteinMPNN scores when compared to a matched subset of natural sequences, indicating that the designed sequences may have higher activity than the natural ones (Fig. 2b). ESM1v is a pLLM developed by Meta Research that was designed for predicting variant effects, ProteinMPNN is a deep learning-based sequence design method that can decode amino acid sequences from structural representations of proteins and score proteins and pLDDT is a metric used by structural prediction tools to evaluate the confidence of predictions. These metrics have previously been used for computational scoring of enzymes13.
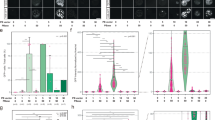
a, Overview of the fine-tuning and sequence generation pipeline. The Progen2-base model was fine-tuned on a set of over 10,000 PiggyBac orthologs identified through the bioprospecting pipeline. Over 100,000 sequences were generated with a sequence identity between 35% and 99% to the HyPB. Sequences were then filtered using a set of basic (gray) and PiggyBac-specific (green) amino acid sequence metrics and scored using a set of scores based on structural (orange) and deep learning (blue) metrics to select a final subset of 22 sequences for experimental validation. b, Distribution of four key metrics (sequence identity, pLDDT, ProteinMPNN score and ESM1v score) for natural sequences from the HyPB cluster at 60% identity (orange) and sequences generated from our progen-ft model (blue) after filtering. The violin plots represent the entire distribution of scores for the two sets of sequences and the internal box plot represents the quartiles for each score, with the center being the median, the bottom and top being the first and third quartiles, respectively, and the whiskers going 1.5× the interquartile range from the top and bottom. Ft, Fourier transform. c, Relative excision for progen-ft-generated variants normalized to HyPB activity (highlighted in green), measured by a transposon excision fluorescence assay. Bars reflect the mean relative excision over the four trials and points represent the mean relative excision of replicates in each trial. Data are presented as the mean values, with n = 5. d, Correlations between calculated and measured features to relative excision of the progen-ft-generated variants. Significant correlations are highlighted in dark blue. Correlation was measured with Pearson’s correlation. e, Targeted integration with top pLLM-generated mutants, measured by a targeted transposon integration GFP reconstitution assay that measures integration of a 1/2 GFP reporter cargo upstream of a stably integrated 2/2 GFP in HEK293T reporter cell line. Triple-mutant (×3) versions of the transposases were made by selecting the residues corresponding to R372A;K375A;D450N in HyPB. Data are presented as the mean values ± 95% CI, with n = 3. f, Targeted transposon integration measured by digital PCR assay in C2C12 mouse myoblast cell lines at TTR and PCSK9 loci for top AI-designed transposases. The sum of integration in both orientations is shown. Data are presented as the mean values ± 95% CI, with n = 2. g, Nontargeted transposon integration measured by fluorescence assay in primary T cells for top bioprospected ortholog Poetur 7 days after electroporation. Data are presented as the mean values ± 95% CI, with n = 2. h, Nontargeted integration of a GFP cargo in primary T cells with HyPB and top synthetic sequences transposases 7 days after cell electroporation. Data are presented as the mean values ± 95% CI, with n = 3.
We experimentally tested 11 sequences from each model (22 total), 15–54 mutations apart from the original HyPB sequence. All of the generated sequences displayed excision activity with an average percentage RFP ranging from 15% to 48% excision (Supplementary Fig. 11c). Of the tested sequences, seven of 22 were significantly more active in excision than the laboratory-evolved HyPB (Fig. 2c) (Mann–Whitney U-test with a P-value cutoff of 0.05). We further evaluated nontargeted integration of the synthetic sequences (Supplementary Fig. 11d). seq3277 was the most active sequence in both excision and nontargeted integration. We termed this sequence Mega-PiggyBac. Curiously, seq136 showed the highest nontargeted integration efficiencies while having baseline excision activities and had the highest number of substitutions (54 amino acids (aa)), most of them in the catalytic region. To evaluate the relevance of the proposed pLLM-based sequence improvement approach, we tested both bioprospected sequences near the Poetur sequence space and single mutants predicted to have improved fitness by ESM1v (‘zero-shot’ approach29) as comparable optimization approaches. In contrast to pLLM, none of these approaches led to mutants with significantly increased nontargeted integration activity (Supplementary Fig. 12).
We gathered multiple metrics to both inform our selection and aid post hoc learning of properties associated with transposase activity. The structural and AI-based scores described above were used to help guide our final selection and, following experimental testing of our variants, certain metrics were found to be correlated to transposase activity. Net charge of the protein, charged fraction of amino acids (ratio of charged amino acids in the sequence) and ProteinMPNN score seemed to be positively correlated with protein activity. In contrast, perplexity scores from the N–>C fine-tuned model, model version (N–>C or C–>N) and Wimley–White30 surface structural similarity scores seemed to be negatively correlated (Fig. 2d and Supplementary Fig. 10a).
We then tested top hits for FiCAT targeted integration (Fig. 2e). We found that synthetic sequence 3277 improved targeted integration twofold, demonstrating that improved pLLM-generated sequences are compatible with programmable gene insertion. We further validated targeted integration with top pLLM-generated sequences in mouse c2c12 myoblast cells at TTR and PCSK9 loci (Fig. 2f). To illustrate the potential impact of bioprospecting guided sequence discovery for therapeutic applications, we stably delivered a GFP transposon cargo with Poetur and AI-designed transposases in T cells, showing higher nontargeted integration for Poetur (Fig. 2g) and for seq136 (Fig. 2h) when compared to HyPB, while seq3277 (Fig. 2h) had same nontargeted integration activity despite having higher excision and targeted integration, underscoring that diversity in pLLM-generated sequences can capture optimization toward different protein properties.
Our work expands the phylogenetic tree of PiggyBac transposons by two orders of magnitude, unveiling a previously unexplored diversity within this family of mobile genetic elements. This expansion led to the discovery and characterization of nine additional active PiggyBac orthologs, broadening the range of transposase variants available for research and biotechnological applications. Among these identified orthologs, two stand out for their exceptional performance, demonstrating activity levels comparable to those of evolved HyPB variants and robust activity in primary T cells, an essential target for many therapeutic applications in gene and cell therapy. Importantly, the discovered orthologs are compatible with the FiCAT programmable gene insertion system. This compatibility paves the way for innovative approaches to gene insertion, enhancing the system’s versatility in applications ranging from gene therapy to synthetic biology. Furthermore, we exemplified how pLLM de novo sequence generation offers a powerful approach to improving transposase activities. This method enhances the optimization process and provides a framework where the modifications are informed by a comprehensive sequence–function relationship. By leveraging the capabilities of pLLM, researchers could use the described method to systematically identify variants with enhanced properties.
Recent work demonstrated substantial activity improvement upon TIR truncation31. Moreover, combining this knowledge on TIR architecture with recently developed genome language models could further improve transposition activity. Additionally, determining how AI-guided activity improvement impacts specificity will be crucial for successfully using these methods for therapeutic protein development.
Our findings underscore the power of combining bioprospection with AI-driven sequence optimization to accelerate the discovery and enhancement of next-generation gene insertion tools. This approach not only expands the PiggyBac toolkit but also provides a valuable framework for the development of additional gene modification tools for precise and efficient genome manipulation applicable across biotechnology and therapeutic fields.
Methods
Retrieval of PiggyBac transposons
Complete PiggyBac transposon sequences were gathered from all available eukaryotic genomes in the NCBI database35 (31,565 genomes) and all PiggyBac elements in the Dfam database (20,638). Dfam sequences were directly downloaded by selecting entries labeled as PiggyBac. NCBI eukaryotic genome-derived transposase sequences were identified using Bath36,37, with a custom hidden Markov model constructed from all active PiggyBac sequences reported in the literature. For NCBI PB retrieval, flanking regions 4 kbp upstream and downstream were included to capture the complete transposon sequence including DNA TIRs. A filter was applied to retain PiggyBac transposases longer than 250 aa. After this filtering, a total of 273,643 PiggyBac were recovered, with a mean transposase length of 500 residues and mean DNA transposon length of 3,298 bp.
To refine the boundaries of each transposon in the NCBI dataset, clustering by RNase H-like domains of the PiggyBac hits at a 0.9 similarity threshold was performed with MMseqs2 (ref. 38), followed by multiple-sequence alignment (MSA) of the complete DNA sequences (including flanking regions) within clusters using MAFFT39. Transposon boundaries were then delimited on the basis of the MSA results.
Filtering for active PiggyBac elements
To identify active PiggyBac transposons from all the transposons identified in the previous step, we applied the following sequential filters:
-
1.
RNase H-like domain identification: The presence of a RNase H-like domain was confirmed using RPS-BLAST40, with the Conserved Domain Database41 as the reference database and selecting only sequences with an RNase H-like domain longer than 250 aa.
-
2.
CRD identification: A total of 50 representative CRDs were manually curated and structurally modeled using AlphaFold3 (ref. 24) to identify residues directly involved in zinc ion coordination. On the basis of this curated set, we derived a set of sequence motifs (Supplementary Table 2), revealing major CRD groups and their variants. CRDs were then identified using regular expressions matching these curated motifs.
-
3.
TIR identification: TIRs were identified in the flanking DNA regions using the EMBOSS tool Palindrome42, focusing on pairs of palindromic sequences located on opposite flanks of the transposon in the first and last 200 bp. We retained only TIRs with at least two palindromic sequences of 10 bp or longer and allowing up to two mismatches. As an additional quality control step, only palindromes in which the two most common nucleotides account for less than 80% of the palindrome were selected.
-
4.
TSD identification: TSDs were searched for with regular expression within the first and last 50 bp of each transposon, using the motif TTAACC, with up to two allowed mismatches.
A total of 116,216 putatively active PiggyBac elements were recovered after applying the filtering process.
Dataset clustering
The filtered dataset was then clustered to reduce redundancy using the RNase H-like domain of the transposase. We performed two clusterings with MMseqs2, one at 0.8 identity and one at 0.6 identity. The 0.8 clustering was performed following transposon annotation 80–80–80 (ref. 43), as it is considered that two transposon elements belong to the same family if they share 80% (or more) sequence identity in at least 80% of their coding or internal domain. This dataset was used for the fine-tuning of the pLLMs. The clustering at 0.6 was performed to make a broader classification of PiggyBac families and used for the phylogenetic analysis. The clustering at 0.8 produced 13,693 clusters, while that at 0.6 produced 2,572 clusters.
Phylogenetic analysis of bioprospected sequences
The phylogenetic tree was built with IQ-TREE (version 1.6.12)44 on the basis of an MSA generated with the 2,572 centroids from the 0.6 clustering with MUSCLE45. Model finder46 was used to select the optimal model for accurate phylogenetic estimation (LG + R10) and UFBoot47 was used for bootstrap approximation with 1,000 replicates. The resulting tree was visualized using iTOL48. Additional PiggyBac domains were identified with RPS-BLAST40. Molecular graphics were generated using UCSF Chimera49.
Blast identification of Poetur orthologs
A search with BLASTn on the core nucleotide database was conducted using Poetur. The whole transposon, including the TIR and TSD were included to find hits that also possessed these motifs. A total of four hits from four different species were manually selected on the basis of them having a coverage higher than 88%, sequence identity higher than 83% and the presence of all necessary functional domains for transposition activity (RNase H-like domain, CRD, TIR and TSD).
Model fine-tuning
The ProGen2-base19 language model of 764 million parameters was fine-tuned on over 13,000 sequences from the PiggyBac orthologs clustered at 0.8. This fine-tuning was performed to give the ProGen2-base model a better understanding of PiggyBac sequences. In this process, the pretrained model was further trained on the PiggyBac orthologs and, as the model trained, the 764 million parameters were updated in a way that aimed to minimize the cross-entropy loss. We fine-tuned two separate models: one model to generate sequences from the N terminus to C terminus and the second to generate sequences from the C terminus to N terminus. Both models were fine-tuned using the full amino acid sequences excluding the N-terminal domain, which was excluded because it is an extremely variable domain. In the HyPB, the N terminus consists of the first 116 aa and, in general, the N terminus is a disordered region leading up to the first double DNA-binding domain region.
The sequences were split using a 80:20 train–test split. In addition to the set of orthologous sequences used in the training, additional wild-type (WT) HyPB sequences (5–10) were added to the training set to bias the model toward HyPB. This allowed us to generate sequences in a closer sequence identity range to HyPB than we were able to without biasing the dataset. Fine-tuning was performed using the Trainer module fetched from Hugging Face over two epochs with a training batch size of 4 and evaluation batch size of 8. A constant learning rate of 5.0 × 10−5 was used and the model was evaluated after every 2,000 steps. Cross-entropy loss was used to evaluate every checkpoint in the model and the checkpoint with the lowest validation loss was used for sequence generation. The remaining Trainer parameters were kept at the default values. A full exploration of the Trainer hyper parameters was not performed as, with these fairly standard parameters, we were able to generate convincing sequences with our desired properties.
AI sequence generation
In both models, 50 aa from WT HyPB were used to prompt sequence generation. An initial prompt was used to give the model enough context to build a PiggyBac-like sequence. In preliminary testing, 50 aa seemed to provide a good balance of giving the models a good starting point without allowing them to replicate the HyPB sequence perfectly. For the N–>C model, the first 50 aa after the N-terminal domain were used and, in the C–>N model, the final 50 aa of the CRD were used to prompt sequence generation. For the C–>N model, sequences were generated ‘backward’ and then reversed to have the standard directionality. The maximum sequence length for both models was set to 500 aa and a temperature of T = 0.5 and nucleus probability P = 0.95 were used.
AI sequence filtering
The generated sequences first went through a set of three basic filters. First, duplicated sequences were removed. Second, sequences with noncanonical amino acids were removed. Third, sequences were filtered using a k-mer repetition filter such that no amino acid motif of six, four, three or two residues was repeated two, three, six or eight times consecutively. The next set of filters were HyPB specific and included testing for a PiggyBac CRD (based on the presence of at least seven cysteine amino acids in the final 50 aa), sequence identity to WT (80–95% to the RNAse H-like and CRD domains) and specific key residues including catalytic site, α-bridge residues, hyperactive residues and another extensive set of key residues including DNA-interacting residues.
For all of these sequences, we calculated perplexity using the ProGen2-base model and the fine-tuned model responsible for generating a given sequence. For a subset of sequences that passed our filters, structures were predicted using ESMFold50. Structures were then compared to the experimentally available PiggyBac structure (PDB 6X67) to extract r.m.s.d. and TM-scores using PyMOL (Schrödinger) and TMAlign51, respectively. Finally, structures were aligned to the experimental PiggyBac structure and several surface properties were calculated using SURFMAP: a tool that projects surface residues from a protein structure into a two-dimensional space and can calculate different amino acid residue properties. The five metrics we calculated using SURFMAP were stickiness, circular variance, Wimley–White, Kyte–Doolittle and electrostatics. We then computed cosine similarities between each surface feature in the generated structures and the experimental structure. Lastly, ProteinMPNN28 and ESM1v29 scores were calculated. ProteinMPNN is a deep learning-based sequence design method that can decode amino acid sequences from structural representations of proteins. ProteinMPNN can also be used to generate a log-likelihood score for any given sequence. Wimley–White is a measure of residue hydrophobicity, which was applied to surface residues in this case using SURFMAP.
An additional set of filters was created to narrow down the final set of sequences. Sequences were required to be in the top 75th percentile for both ProteinMPNN and ESM1v scores, sequences were filtered on length to exclude sequences that were too short, a conservative pLDDT filter of 90 was used and an acceptable range for net charge of the proteins was established. After this, sequences were selected manually in an attempt to cover sequence identities in the range of 90–97% to the entire HyPB sequence with high-quality sequences. During this manual selection process, sequences with a higher proportion of the key residues were selected for and any sequences that had particularly bad scores in any of the calculated metrics were avoided. A final selection of 22 sequences was made.
In silico deep mutational scan
ESM1v was used in a zero-shot version where the Poecliopsis amino acid sequence was given as an input. ESM1v creates a fitness score for all possible amino acids for residue position by calculating a log odds ratio, assuming an additive model when multiple substitutions exist. Then, the sum is made over the substituted positions and the sequence is masked at every substituted position29.
Variant prediction was run in Google Colab Pro with one A-100 GPU with 80 GB of RAM. The script used to run the variant prediction can be found on GitHub (https://github.com/Alejo945/IS-HyPB). The output is a TSV file with all possible variants and their scores.
Plasmid DNA sequences
Transposase ORF amino acid sequences were codon-optimized for Homo sapiens and ordered and synthesized as gene fragments to TWIST biosciences. Gene fragments were cloned into a cytomegalovirus-based expression vector by Golden Gate assembly using Esp3I restriction enzyme. Transposon (cargo vector) plasmid sequences were defined as the first 150 bp from the transposon ends from both 5′ and 3′ TIR sequences and synthesized as gene fragments by TWIST biosciences with added overhangs for golden gate assembly. An EF1α RFP poly(A) expression cassette was included between the TIR. Triple mutant (×3, R372A;K375A;D450N in Trichoplusia ni) residue selection was performed by aligning the ortholog sequences to the T. ni PiggyBac mutated sequence. All plasmid sequences are available in Supplementary Table 1.
Cell culture
Hek293T cells (Invitrogen, R70007), were cultured in DMEM supplemented with high glucose (Gibco, Thermo Fisher), 10% FBS, 2 mM glutamine, 100 U per ml penicillin and 0.1 mg ml−1 streptomycin at 37 °C in a 5% CO2 incubator.
PCR excision activity assay
To detect excision in bioprospected transposases, 120,000 cells were seeded per adherent p24 well 1 day before transfection. Plasmid DNA was mixed at a 1:3 ratio of transposase and RFP transposon, with 0.035 pmol of transposase used per p24 well plate. Then, 48 h after transfection, cells were collected and plasmid extraction was performed using an NZYMiniprep kit (NZYtech, MB01001). TIR-flanking primers (Supplementary Table 4) were used to detect transposon excision. The 2,900-bp and 1,200-bp bands indicated nonexcised and excised transposon, respectively.
Nontargeted transposon integration fluorescence assay
To evaluate stable transposon integration activity, 120,000 cells were seeded per adherent p24 well a day before transfection. Plasmid DNA was mixed with and RFP transposon at a ratio of 1:3:5, with 0.035 pmol of transposase used per p24 well plate. For transfection experiments, cells were transfected with polyethyleneimine (PEI, Thermo Fisher Scientific) at a 1:3 ratio of DNA and PEI in Opti-MEM. RFP expression of the transposon cargo vector was assessed 2 days and 20 days after transfection using cell cytometry with the Cytek Aurora CS system. The RFP signal at day 20 was considered indicative of stable transgene integration.
Transposon excision fluorescence assay
To quantify the excision activity of AI-generated transposases, a fluorescent excision reporter system was used. HEK293T cells were seeded in 24-well plates at a density of 120,000 cells per well 24 h before transfection to ensure approximately 70% confluency on the day of transfection. Transfections were performed in 24-well plates using PEI (Thermo Fisher Scientific) at a 1:3 ratio of DNA and PEI in Opti-MEM (Thermo Fisher). Transposase-expressing plasmid was cotransfected with plasmid containing a disrupted mCherry reporter sequence flanked by transposase recognition sites, leading to mCherry restoration upon excision (Supplementary Fig. 6). Transposase and transposon plasmids were mixed at a 1:3 ratio, with a total of 0.035 pmol of transposase. Then, 72 h after transfection, cells were collected and mCherry reporter expression was assessed by flow cytometry using the Cytek Aurora CS system.
Targeted transposon integration digital PCR assay
To quantify targeted integration of AI-generated transposases in the FiCAT system, C2C12 cells (American Type Cell Collection, CRL-1772) were cultured in DMEM (Gibco, Thermo Fisher) supplemented with 10% FBS, 2 mM l-glutamine, 100 U per ml penicillin and 0.1 mg ml−1 streptomycin. Cells were maintained in a 37 °C incubator with 5% CO2. Electroporation was conducted using the E Cell Line 4D-Nucleofector X Kit S (Lonza). On the day of electroporation, cells were washed with PBS, detached using trypsin–EDTA (Gibco) and adjusted to a concentration of 2 × 105 cells per condition. The cell suspension was prepared in 20 µl of nucleofection master mix buffer, consisting of 16.4 µl Nucleofector solution and 3.6 µl of supplement 1 (Lonza). Subsequently, each condition was conucleofected with a DNA plasmid encoding the triple-mutant variants (PB×3), Cas9, different guide RNAs (gRNAs) and transposon plasmids in a 1:1:3:3 molar ratio, using a maximum of 10% of the final sample volume. Lastly, each condition was transferred into Nucleocuvette vessels and electroporation was carried out using the CD-137 program. After electroporation, 100 µl of prewarmed complete medium was added and cells were carefully resuspended and transferred into a 24-well plate containing 500 µl of complete medium for recovery and expansion. Then, 4 days after electroporation, the cells were processed as follows: (1) one third were collected for genomic extraction; (2) one third were analyzed for GFP reporter expression by flow cytometry using the Cytek Aurora CS system; and (3) one third were maintained in culture until episomal disappearance. Genomic extraction was performed using Qiagen DNeasy blood and tissue kit. Primers and probes were obtained from PrimeTime qPCR probes (Integrated DNA Technologies). The assay was designed using an endogenous control and evaluating the junction PCR for both integration orientations. Reaction mixtures (44 μl) were prepared containing QIAcuityDx Universal master mix (1×), MgCl2 (6.28 mM), primers (0.73 µM), probes (0.63 µM), a restriction enzyme (0.25 U per µL) and 12.5 ng of sample DNA. These mixtures were loaded onto a QIAcuityDx Nanoplate 26k 24-well (260001) for quantification, following the preparation protocol provided in the QIAcuityDx Universal master mix kit (260102). Thermal cycling protocol consisted of an initial enzyme activation step at 95 °C for 2 min, followed by 40 cycles of a two-step amplification: denaturation at 95 °C for 15 s and annealing and extension at 60 °C for 30 s. For digital PCR analysis, the absolute DNA quantification per sample (copies per genome) was determined using QIAcuity Software. Primer sequences are described in Supplementary Table 6.
Targeted transposon integration fluorescence and qPCR assay
To quantify targeted integration of bioprospected transposases in the FiCAT system, Plasmids encoding the triple-mutant variants (PB×3) were cotransfected with Cas9, gRNA AAVS1-3, transposase and transposon plasmids at a 1:1:3:5 molar ratio in 0.5 M Hek23T cells seeded in a p6 plate the day before transfection. Cells were analyzed for RFP expression 2 days after transfection to estimate transfection efficiency using cell cytometry with the Cytek Aurora CS system. Cells were maintained in culture to measure overall integration levels after 3 weeks. In parallel, to enrich cells for junction qPCR, two rounds of enrichment by GFP sorting were conducted with BD FACSAria (Biosciences), 1 week and 2 weeks after transfection. Genomic DNA was extracted using Quiagen DNeasy blood and tissue kit column 4 days after the second sorting. A 3′ junction PCR was performed and sequenced on an Illumina MiSeq Nano kit 500 cycles (v2). A 3′ junction qPCR was performed to compare targeted integration across bioprospected transposases.
Targeted transposon integration GFP reconstitution assay
To quantify targeted integration in AI-generated PiggyBac transposases in the FiCAT system, a previously described GFP reconstitution assay52 was used. For GFP targeted integration assays, a reporter HEK293T cell line containing genomically integrated 2/2 GFP was transfected using a 1/2 GFP encoding transposon (Supplementary Fig. 6). A total of 240,000 2/2 GFP HEK293T reporter cells were seeded in a 12-well plate 1 day before transfection. Cells were transfected with Lipofectamine 3000 (Invitrogen, L3000001) using Cas9, 2/2 GFP-targeting gRNA, transposase and transposon plasmids at a 1:1:3:5 molar ratio. Cells were analyzed for GFP expression 5 days after transfection to estimate targeted integration efficiency using cell cytometry with the Cytek Aurora CS system. The 2/2 GFP was integrated using the Sleeping Beauty (SB100x) transposase system53. Reporter DNA sequences are available in supplementary Table 3.
Nontargeted transposon integration fluorescence assay in T cells
To assess nontargeted integration of the PiggyBac and AI-generated orthologs in T cells, peripheral blood mononuclear cells from two different donors, isolated from buffy coats and cryopreserved, were thawed and seeded on p24-coated plates containing anti-CD3/CD28 (1:1,000; BD Sciences) at a density of 1 × 106 cells per ml in 3 ml of CTS OpTmizer T cell expansion SFM medium (Thermo Fisher), supplemented with interleukin (IL)-7 and IL-15 (10 ng ml−1 each; Miltenyi Biotec). Buffy coats were obtained from the Barcelona Blood and Tissue Bank upon institutional review board approval.
For nontargeted integration in bioprospected orthologs, on the third day of culture, electroporation was conducted using the P3 primary cell 4D-Nucleofector X kit (Lonza). Cells were washed with PBS (Capricorn) and adjusted to a concentration of 7.5 × 105 cells per condition. The cell suspension was prepared in 20 µl of nucleofection buffer, consisting of 16.4 µl of P3 primary cell Nucleofector solution and 3.6 µl of supplement 1 (Lonza). Subsequently, 1 µg of each DNA plasmid was added to the suspension and electroporation was carried out using the EO-115 nucleofection program. The minimal backbone GenCircle-TIR_CAR19-GFP transposon plasmid was used (GenCircle, manufactured by Genscript). For each evaluated transposase, conditions with transposase + transposon and transposon only were electroporated in duplicates to differentiate between episomal and integrated signals. Following electroporation, 80 µl of complete medium was added and cells were incubated at 37 °C for 20 min. The cells were then carefully resuspended and transferred to a fresh p24 plate containing 500 µl of medium for recovery and expansion. Approximately one third of the well volume was used for flow cytometric analysis using the Aurora system (Cytek) to assess RFP expression levels at 4 and 7 days after transfection.
For nontargeted integration of AI-generated orthologs, On the third day of culture, electroporation was conducted using the P3 primary cell 4D-Nucleofector X kit (Lonza). Cells were washed with PBS (Capricorn) and adjusted to a concentration of 1 × 106 cells per condition. The cell suspension was prepared in 20 µl of nucleofection buffer, consisting of 16.4 µl of P3 primary cell Nucleofector solution and 3.6 µl of supplement 1 (Lonza). Subsequently, 1 µg of each DNA plasmid was added to the suspension and electroporation was carried out using the EH-115 nucleofection program. The minimal backbone GenCircle-TIR_CAR19-GFP transposon plasmid was used (GenCircle, manufactured by Genscript).For each evaluated transposase, conditions with transposase + transposon and transposon only were electroporated in duplicates to differentiate between episomal and integrated signals. Following electroporation, 80 µl of complete medium was added and cells were incubated at 37 °C for 20 min. The cells were then carefully resuspended and transferred to a fresh p24 plate containing 500 µl of medium for recovery and expansion. Medium supplemented with H-151 (MedChemExpress, HY-112693) STING inhibitor at 2 µM was added. Approximately one third of the well volume was used for flow cytometric analysis using the Aurora system (Cytek) to assess GFP expression levels at 4 and 7 days after transfection.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Experimentally tested transposon sequence files are available in Supplementary Table 1. Top active transposon and transposase plasmids were deposited to Addgene.
Code availability
Model fine-tuning and PiggyBac generation code is available from Github (https://github.com/Integra-tx/Piggybac_bioprospecting_pipeline).
References
Wang, J. Y. & Doudna, J. A. CRISPR technology: a decade of genome editing is only the beginning. Science 379, eadd8643 (2023).
Yarnall, M. T. N. et al. Drag-and-drop genome insertion of large sequences without double-strand DNA cleavage using CRISPR-directed integrases. Nat. Biotechnol. 41, 500–512 (2023).
Mukhametzyanova, L. et al. Activation of recombinases at specific DNA loci by zinc-finger domain insertions. Nat. Biotechnol. 42, 1844–1854 (2024).
Li, X. et al. PiggyBac transposase tools for genome engineering. Proc. Natl Acad. Sci. 110, E2279–E2287 (2013).
Yusa, K., Zhou, L., Li, M. A., Bradley, A. & Craig, N. L. A hyperactive PiggyBac transposase for mammalian applications. Proc. Natl Acad. Sci. USA 108, 1531–1536 (2011).
Mitra, R. et al. Functional characterization of PiggyBat from the bat Myotis lucifugus unveils an active mammalian DNA transposon. Proc. Natl Acad. Sci. USA 110, 234–239 (2013).
Chen, Q. et al. Structural basis of seamless excision and specific targeting by PiggyBac transposase. Nat. Commun. 11, 3446 (2020).
Yuan, Y.-W. & Wessler, S. R. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl Acad. Sci. USA 108, 7884–7889 (2011).
Guo, M. et al. PiggyBac transposon mining in the small genomes of animals. Biology 13, 24 (2024).
Ruffolo, J. A. et al. Design of highly functional genome editors by modeling the universe of CRISPR–Cas sequences. Nature https://doi.org/10.1038/s41586-025-09298-z (2025).
Hayes, T. et al. Simulating 500 million years of evolution with a language model. Science 387, 850–858 (2025).
Alley, E. C., Khimulya, G., Biswas, S., AlQuraishi, M. & Church, G. M. Unified rational protein engineering with sequence-based deep representation learning. Nat. Methods 16, 1315–1322 (2019).
Watson, J. L. et al. De novo design of protein structure and function with RFdiffusion. Nature 620, 1089–1100 (2023).
Lauko, A. et al. Computational design of serine hydrolases. Science 388, eadu2454 (2025).
Galvan, D. L. et al. Genome-wide mapping of PiggyBac transposon integrations in primary human T cells. J. Immunother. 32, 837–844 (2009).
Luo, W. et al. Comparative analysis of chimeric ZFP-, TALE- and Cas9-PiggyBac transposases for integration into a single locus in human cells. Nucleic Acids Res. 45, 8411–8422 (2017).
Hew, B. E., Sato, R., Mauro, D., Stoytchev, I. & Owens, J. B. RNA-guided PiggyBac transposition in human cells. Synth. Biol. 4, ysz018 (2019).
Adrian, K. et al. RNA-guided retargeting of Sleeping Beauty transposition in human cells. eLife 9, e53868 (2020).
Nijkamp, E., Ruffolo, J. A., Weinstein, E. N., Naik, N. & Madani, A. Progen2: exploring the boundaries of protein language models. Cell Syst. 14, 968–978 (2023).
Storer, J., Hubley, R., Rosen, J., Wheeler, T. J. & Smit, A. F. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mobile DNA 12, 2 (2021).
Bouallègue, M., Rouault, J.-D., Hua-Van, A., Makni, M. & Capy, P. Molecular evolution of PiggyBac superfamily: from selfishness to domestication. Genome Biol. Evol. 9, 323–339 (2017).
Cosby, R. L. et al. Recurrent evolution of vertebrate transcription factors by transposase capture. Science 371, eabc6405 (2021).
Zhang, H.-H., Peccoud, J., Xu, M.-R.-X., Zhang, X.-G. & Gilbert, C. Horizontal transfer and evolution of transposable elements in vertebrates. Nat. Commun. 11, 1362 (2020).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Yusa, K. PiggyBac transposon. Microbiology Spectrum 3, MDNA3–0028–2014 (2015).
Schweke, H., Mucchielli, M.-H., Chevrollier, N., Gosset, S. & Lopes, A. SURFMAP: a software for mapping in two dimensions protein surface features. J. Chem. Inf. Model. 62, 4211–4219 (2022).
Sanner, M. F., Olson, A. J. & Spehner, J.-C. Reduced Surface: an efficient way to compute molecular surfaces. Biopolymers 38, 305–320 (1996).
Dauparas, J. et al. Robust deep learning-based protein sequence design using ProteinMPNN. Science 378, 49–56 (2022).
Meier, J. et al. Language models enable zero-shot prediction of the effects of mutations on protein function. Preprint at bioRxiv https://doi.org/10.1101/2021.07.09.450648 (2021).
Wimley, W. C. & White, S. H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3, 842–848 (1996).
Hickman, A. B. et al. Activity of the mammalian DNA transposon PiggyBat from Myotis lucifugus is restricted by its own transposon ends. Nat. Commun. 16, 458 (2025).
Tian, J. et al. Mage transposon: a novel gene delivery system for mammalian cells. Nucleic Acids Res. 52, 2724–2739 (2024).
Wu, C. & Wang, S. PLE-wu, a new member of PiggyBac transposon family from insect, is active in mammalian cells. J. Biosci. Bioeng. 118, 359–366 (2014).
Lindstrom-Vautrin, J. & Agudelo, A. PiggyBac bioprospecting pipeline. GitHub https://github.com/Integra-tx/Piggybac_bioprospecting_pipeline.git (2025).
Kitts, P. A. et al. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res. 44, D73–D80 (2016).
Eddy, S. R. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 (2011).
Krause, G. R., Shands, W. & Wheeler, T. J.Sensitive and error-tolerant annotation of protein-coding DNA with BATH. Bioinform. Adv. 4, vbae088 (2024).
Steinegger, M. & Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
Lu, S. et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268 (2020).
Rice, P., Longden, I. & Bleasby, A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 (2000).
Wicker, T. et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982 (2007).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Edgar, R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Zhang, Y. & Skolnick, J. TM-align: a protein structure alignment algorithm based on TM-score. Nucleic Acids Res. 33, 2302–2309 (2005).
Pallarès-Masmitjà, M. et al. Find and cut-and-transfer (FiCAT) mammalian genome engineering. Nat. Commun. 12, 7071 (2021).
Mátés, L. et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 41, 753–761 (2009).
Acknowledgements
We thank C. Feschotte for feedback on the transposon bioprospecting pipeline, G. M. Church for advice on approaches for machine learning, AI and directed evolution and M. Artigues for her help in designing the initial filtering process for the Progen2 generated sequences. Integra Therapeutics received funding from NEOTEC (CDTI, SNEO-20222363). Universitat Pompeu Fabra received funding from UPGRADE (European Union Horizon 2020, grant agreement no. 825825) and Ministerio de Economia, Industria y Competitividad de España (Plan Estatal 2013-2016 (grant agreement no. PID2020-118597RB-I00).
Author information
Authors and Affiliations
Contributions
D.I., A.S.M. and M. Güell conceptualized the study. A.A. and D.I. designed the bioprospecting pipeline. A.A. implemented the bioprospecting pipeline. J.L.-V. implemented the LLM and fine-tuning work with help from N.F., A.A. and D.I. D.I. and J.J.-W. designed the experiments with help from R.D. and M. Gallo. M. Gallo, R.D. and J.J.W. performed the cell experiments. I.H. assisted with the sequence assembly. A.R. and P.P. performed the T cell work. F.B. contributed to genome data accession and zero-shot modeling. J.H.-V. performed insertional profiling and molecular characterization of editing and transposition outcomes. M.S.-G. analyzed the targeted integration data. D.I. and M. Güell supervised the study. A.A., D.I. and J.V.L. plotted the data. A.A., D.I., M. Güell and J.L.V. wrote the paper with contributions from all authors.
Corresponding authors
Ethics declarations
Competing interests
A.A., J.L.-V., J.J.-W., M. Gallo, M.S.-G., R.D., M. Güell, A.S.-M., N.F. and D.I. are employed or have consulted for Integra Therapeutics. M. Güell and A.S.-M. are shareholders of Integra therapeutics. D.I., M. Güell, A.S.-M., A.A. and R.D. have filed a patent application (US Patent application no. 63/505485) related to this work.
Peer review
Peer review information
Nature Biotechnology thanks Zoltán Ivics, Jesse Owens and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–12.
Supplementary Tables
Supplementary Tables 1–7.
Supplementary Data
Unprocessed gel images for Supplementary Fig. 6.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ivančić, D., Agudelo, A., Lindstrom-Vautrin, J. et al. Discovery and protein language model-guided design of hyperactive transposases. Nat Biotechnol (2025). https://doi.org/10.1038/s41587-025-02816-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41587-025-02816-4