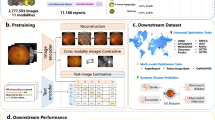
Abstract
In the context of an increasing need for clinical assessments of foundation models, we developed EyeFM, a multimodal vision–language eyecare copilot, and conducted a multifaceted evaluation, including retrospective validations, multicountry efficacy validation as a clinical copilot and a double-masked randomized controlled trial (RCT). EyeFM was pretrained on 14.5 million ocular images from five imaging modalities paired with clinical texts from global, multiethnic datasets. Efficacy validation invited 44 ophthalmologists across North America, Europe, Asia and Africa in primary and specialty care settings, highlighting its utility as a clinical copilot. The RCT—a parallel, single-center, double-masked study—assessed EyeFM as a clinical copilot in retinal disease screening among a high-risk population in China. A total of 668 participants (mean age 57.5 years, 79.5% male) were randomized to 16 ophthalmologists, equally allocated into intervention (with EyeFM copilot) and control (standard care) groups. The primary endpoint indicated that ophthalmologists with EyeFM copilot achieved higher correct diagnostic rate (92.2% versus 75.4%, P < 0.001) and referral rate (92.2% versus 80.5%, P < 0.001). Secondary outcome indicated improved standardization score of clinical reports (median 33 versus 37, P < 0.001). Participant satisfaction with the screening was similar between groups, whereas the intervention group demonstrated higher compliance with self-management (70.1% versus 49.1%, P < 0.001) and referral suggestions (33.7% versus 20.2%, P < 0.001) at follow-up. Post-deployment evaluations indicated strong user acceptance. Our study provided evidence that implementing EyeFM copilot can improve the performance of ophthalmologists and the outcome of patients. Chinese Clinical Trial Registry registration: ChiCTR2500095518.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
For the reproduction of our algorithm code, we have also deposited a minimum dataset (https://zenodo.org/records/15546254; ref. 41), which is publicly available for scientific research and non-commercial use. The data supporting the findings of this trial are available within the paper and its supplementary information files. All requests for further data sharing will be reviewed by the data management committees from participating institutions and by the ethics committee of Shanghai Health and Medical Centre, China, to verify whether the request is subject to any intellectual property or confidentiality obligations and will be accessible with informed consents. Requests for access to deidentified individual-level data from this trial can be submitted via email to B.S. (shengbin@sjtu.edu.cn) with detailed proposals for approval and will be evaluated on a case-by-case basis and responded to within 60 days. Investigators who consent to the terms of the data transfer agreement, including, but not limited to, the use of these data only for academic purposes and to protect the confidentiality of the data and limit the possibility of identification of patients, will be granted access. Source data are provided with this paper.
Code availability
The code being used in the current study for developing the algorithm is provided at https://github.com/eyefm/EyeFM.
References
Zhou, Y. et al. A foundation model for generalizable disease detection from retinal images. Nature 622, 156–163 (2023).
Chen, R. J. et al. Towards a general-purpose foundation model for computational pathology. Nat. Med. 30, 850–862 (2024).
Lu, M. Y. et al. A visual-language foundation model for computational pathology. Nat. Med. 30, 863–874 (2024).
Topol, E. J. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 25, 44–56 (2019).
Tanno, R. et al. Collaboration between clinicians and vision–language models in radiology report generation. Nat. Med. 31, 599–608 (2025).
Norden, J, G. & Shah, N. R. What AI in health care can learn from the long road to autonomous vehicles. NEJM Catalyst https://catalyst.nejm.org/doi/abs/10.1056/CAT.21.0458 (2022).
You, J. G., Hernandez-Boussard, T., Pfeffer, M. A., Landman, A. & Mishuris, R. G. Clinical trials informed framework for real world clinical implementation and deployment of artificial intelligence applications. NPJ Digit. Med. 8, 107 (2025).
Longhurst, C. A., Singh, K., Chopra, A., Atreja, A. & Brownstein, J. S. A call for artificial intelligence implementation science centers to evaluate clinical effectiveness. NEJM AI https://doi.org/10.1056/AIp2400223 (2024).
Gupta, A., Savarese, S., Ganguli, S. & Fei-Fei, L. Embodied intelligence via learning and evolution. Nat. Commun. 12, 5721 (2021).
Colunga-Lozano, L. E. et al. Clinical judgment shows similar and sometimes superior discrimination compared to prognostic clinical prediction models: a systematic review. J. Clin. Epidemiol. 165, 111200 (2024).
Bommasani, R. et al. On the opportunities and risks of foundation models. Preprint at https://arxiv.org/abs/2108.07258 (2022).
Howell, M. D., Corrado, G. S. & DeSalvo, K. B. Three epochs of artificial intelligence in health care. JAMA 331, 242–244 (2024).
Lu, M. Y. et al. A multimodal generative AI copilot for human pathology. Nature 634, 466–473 (2024).
Future of Health: The Emerging Landscape of Augmented Intelligence in Health Care. https://www.ama-assn.org/system/files/future-health-augmented-intelligence-health-care.pdf (American Medical Association, 2024).
Yang, J. et al. Generalizability assessment of AI models across hospitals in a low-middle and high income country. Nat. Commun. 15, 8270 (2024).
Avram, O. et al. Accurate prediction of disease-risk factors from volumetric medical scans by a deep vision model pre-trained with 2D scans. Nat. Biomed. Eng. 9, 507–520 (2024).
Street, A., Kersaudy Kerhoas, M. & Ndlovu, Z. From equitable access to equitable innovation: rethinking bioengineering for global health. Nat. Rev. Bioeng. 2, 444–446 (2024).
Matheny, M. E., Whicher, D. & Thadaney Israni, S. Artificial intelligence in health care: a report from the National Academy of Medicine. JAMA 323, 509–510 (2020).
van de Sande, D. et al. To warrant clinical adoption AI models require a multi-faceted implementation evaluation. NPJ Digit. Med. 7, 58 (2024).
Hadziahmetovic, M., Nicholas, P., Jindal, S., Mettu, P. S. & Cousins, S. W. Evaluation of a remote diagnosis imaging model vs dilated eye examination in referable macular degeneration. JAMA Ophthalmol. 137, 802–808 (2019).
Liu, H., Li, C., Wu, Q. & Lee, Y. J. Visual instruction tuning. In Advances in Neural Information Processing Systems 36 (eds Oh, A. et al.) https://papers.nips.cc/paper_files/paper/2023/file/6dcf277ea32ce3288914faf369fe6de0-Paper-Conference.pdf (Curran Associates, 2023).
Touvron, H. et al. Llama 2: open foundation and fine-tuned chat models. Preprint at https://arxiv.org/abs/2307.09288 (2023).
Bachmann, R., Mizrahi, D., Atanov, A. & Zamir, A. MultiMAE: Multi-modal Multi-task Masked Autoencoders. In Computer Vision – ECCV 2022 (eds Avidan, S. et al.) 348–367 (Springer-Verlag, 2022).
Rafailov, R. et al. Direct preference optimization: your language model is secretly a reward model. In Proc. of the 37th International Conference on Neural Information Processing Systems (eds Oh, A. et al.) 53728–53741 (Curran Associates, 2023).
McMahan, B., Moore, E., Ramage, D., Hampson, S. & Arcas, B. A. Y. Communication-efficient learning of deep networks from decentralized data. In Proc. of the 20th International Conference on Artificial Intelligence and Statistics (eds Singh, A. & Zhu, J.) 1273–1282 (PMLR, 2017).
Chen, X. et al. FFA-GPT: an automated pipeline for fundus fluorescein angiography interpretation and question-answer. NPJ Digit. Med. 7, 111 (2024).
Singhal, K. et al. Toward expert-level medical question answering with large language models. Nat. Med. 31, 943–950 (2025).
McDuff, D. et al. Towards accurate differential diagnosis with large language models. Nature 642, 451–457 (2025).
Ting, D. S. W. et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA 318, 2211–2223 (2017).
Wang, W. et al. Learning two-stream CNN for multi-modal age-related macular degeneration categorization. IEEE J. Biomed. Health Inform. 26, 4111–4122 (2022).
He, M. et al. Prevalence and clinical characteristics of glaucoma in adult Chinese: a population-based study in Liwan District, Guangzhou. Invest. Opthalmol. Vis. Sci. 47, 2782–2788 (2006).
Liu, X. et al. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat. Med. 26, 1364–1374 (2020).
Bourne, R. et al. Trends in prevalence of blindness and distance and near vision impairment over 30 years: an analysis for the Global Burden of Disease Study. Lancet Glob. Health 9, e130–e143 (2021).
Trott, M. et al. Eye disease and mortality, cognition, disease, and modifiable risk factors: an umbrella review of meta-analyses of observational studies. Eye 36, 369–378 (2022).
Xiong, K., Mao, H., Zhang, Q., Lei, C. & Liang, Y. Associations between vision impairment and multimorbidity among older Chinese adults: results from the China health and retirement longitudinal study. BMC Geriatr. 23, 688 (2023).
Zheng, D. D. et al. Patterns of chronic conditions and their association with visual impairment and health care use. JAMA Ophthalmol. 138, 387–394 (2020).
Holden, B. A. et al. Global prevalence of myopia and high myopia and temporal trends from 2000 through 2050. Ophthalmology 123, 1036–1042 (2016).
Singhal, K. et al. Large language models encode clinical knowledge. Nature 620, 172–180 (2023).
Lewis, J. R. & Sauro, J. in Human Centered Design (ed Kurosu, M.) 94–103 (Springer, 2009).
Hillis, S. L. & Soh, B. P. Obuchowski-Rockette analysis for multi-reader multi-case (MRMC) readers-nested-in-test study design with unequal numbers of readers. Proc. SPIE Int. Soc. Opt. Eng. 12467, 124670F (2023).
EyeFM study group EyeFM sample dataset. Zenodo https://zenodo.org/records/15546254 (2025).
Acknowledgements
We thank H. Li and Z. Li for creating the illustrations and icons. This study was supported by the National Key R&D Program of China (2022YFC2502800), the National Natural Science Foundation of China (82388101) and the Beijing Natural Science Foundation (IS23096) to T.Y.W.; the National Natural Science Foundation of China (62272298), the Noncommunicable Chronic Diseases-National Science and Technology Major Project (2023ZD0509202 & 2023ZD0509201), the National Key Research and Development Program of China (2022YFC2407000) to B.S.; and the Noncommunicable Chronic Diseases-National Science and Technology Major Project (2023ZD0509202 & 2023ZD0509201), the Clinical Special Program of Shanghai Municipal Health Commission (20224044) and the Three-Year Action Plan to Strengthen the Construction of the Public Health System in Shanghai (2023-2025 GWVI-11.1-28) to T.C. These funders/sponsors had no role in the design or conduct of the study.
Author information
Authors and Affiliations
Consortia
Contributions
T.Y.W. and B.S. conceived and supervised the project. T.Y.W., B.S., Yilan Wu, Z.G., D.Z. and Y.F.Z. designed the study. B. Qian, Y.Q. and P.Z. designed the deep learning algorithm and the computational framework. T.Y.W., Yilan Wu, B. Qian, T.L, Y.Q., Z.G., D.Z. and Y.F.Z. contributed to the initial drafting of the manuscript. Y.J., P.Z., Y.Z., Q.P., C.Y., J.S., A.G., M.G.-B., M.G., A.S., W.S., L.Z. and You Wu helped with data collection. S.M., R.R., B.S.T., J.A.O.Ñ., T.A.K., H.L., Y.J., A.R.R., D.Y., Z.M., D.W., Y.C., W.Y., R.D., X. Zhao, C.Z., X.W., Y.C., Q.W., H.X., S.K.H.S., J.Y.Y.C., V.T.T.C., H.-T.X., R.W., J.L., Shan Lin, Z.X., N.G., J.E., A.L., F.D., MA., P.C., T.A.M., Y.H., Y.Z., Shiqun Lin, X.B., J.W., X.Y., H.Z., Y.L, B. Qu, H.Y., M.G., M.Z., W.S., L.M.S., F.P., B.S.S., A.A.T., C.E.N.M., P.V., D.S., A.K.T., D.B., U.K., A.K., T.I., P.L.P.W., M.J.A., N.N.A. and I.E.-T. participated in prospective validations. T.C., X. Zhang, Y.H., X.B., J.W., X.Y., H.Z. and Y.L. conducted the data collection and analysis in the RCT. J.G., P.R., S.S., P.A.K., L.-L.L., C.Y.C., G.S.W.T., Y.X.W., Y.-C.T., C.-Y.C., Y.F.Z., B.S. and T.Y.W. contributed to collaboration organization and provided critical revision of the manuscript for important intellectual content. All authors provided critical comments and reviewed the manuscript. All authors discussed the results and approved the final version before submission.
Corresponding authors
Ethics declarations
Competing interests
Y.J. is a patent holder of Optovue/Visionix, Inc., Optos plc and Genentech, Inc. She receives financial support from Genentech, Inc., and she receives financial compensation from Optovue/Visionix, Inc. and Genentech, Inc. P.A.K. is a co-founder of Cascader Ltd., has acted as a consultant for Retina Consultants of America, Roche, Boehringer Ingleheim and Bitfount and is an equity owner in Big Picture Medical. He has received speaker fees from Zeiss, Thea, Apellis and Roche. He has received travel support from Bayer and Roche. He has attended advisory boards for Topcon, Bayer, Boehringer Ingleheim and Roche. T.Y.W. is a consultant for AbbVie Pte Ltd., Aldropika Therapeutics, Bayer, Boehringer Ingelheim, Zeiss, Genentech, Iveric Bio, Novartis, Opthea Limited, Plano, Quaerite Biopharm Research Ltd., Roche, Sanofi and Shanghai Henlius. He is an inventor, holds patents and is a co-founder of start-up companies EyRiS and Visre, which have interests in, and develop digital solutions for, eye diseases. All potential conflicts of interests for consultancy, advisory boards and positions in the start-up companies and financial renumeration, if any, are managed by institutional policies under SingHealth and Tsinghua University. The other authors declare no competing interests.
Peer review
Peer review information
Nature Medicine thanks Tae Keun Yoo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Lorenzo Righetto, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Envision the use case of EyeFM as a whole workflow copilot for eyecare under different clinical settings.
For patients attending ocular disease screening in the primary care centre where only low-cost examination technics are available, EyeFM can use its single modality and cross-modality ability to assist disease detection, followed with screening report writing assisted by its vision-question answering ability. Then, some patients will be referred to specialty care settings for further examination, where EyeFM can use its integrated modality disease detection or even zero-shot ability to facilitate further diagnosis. The image-report writing ability and vision-question answering can also assist to improve the efficiency of clinical report and patient letter drafting in specialty care settings.
Extended Data Fig. 2 The structural diagram of human-knowledge encoding in pretraining and application phase for EyeFM.
a) The diagram of pretraining process for EyeFM. EyeFM was first pretrained for its image encoder with five modalities of images, then conducted vision-language joint pretraining. The image module includes one encoder and five decoders. The encoder comprises 24 Transformer blocks. Each decoder comprises two Transformer blocks. The linear projection layer is implemented with a single convolutional layer. In the vision-language module, the projection is implemented using a single linear layer, which serves to connect the image encoder with the language module. The language module is based on LLaMA 2 architecture with 7 billion parameters. b) The diagram of human-in-the-loop process for EyeFM. EyeFM human-in-the-loop utilized DPO and federated learning for distributed knowledge evolution.
Extended Data Fig. 3 Glossary table and the clinical tasks related with each validation experiment.
First, we conducted retrospective validations, comparing EyeFM with prior benchmarks of medical foundation models. This step serves as the foundation for evaluating the model’s performance and safety when progressing to clinical applications. Second, we conducted reader studies and real-world study prospectively to test the efficiency of EyeFM as a clinical copilot to assist ophthalmologists. This step bridged the gap between the performance of the model its own and its efficiency when applicated by clinicians. At last, we validated EyeFM with a Randomised Controlled Trial (RCT). Colored chart, above, validation experiments and their corresponding relationship with the functions of EyeFM. The x-axis represents different tasks of EyeFM and y-axis represents validation experiments. Cells were coloured if the experiments have validated corresponding functions. Below, glossary table of the tasks, clinical scenario and experiments. RCT, randomised controlled trial.
Extended Data Fig. 4 Experiment 1 – Retrospective validation of EyeFM on multi-ethnic datasets.
a) For disease detection on CFP, the sample sizes and P values are: DR (n = 1501, P = 0.042), glaucoma suspect (n = 405, P = 0.533), AMD suspect (n = 370, P = 0.627), MMD (n = 643, P = 0.030). For disease detection on OCT, the sample sizes and P values are: ciDME (n = 523, P = 0.002), glaucoma (n = 412, P = 0.333), AMD (n = 379, n = 0.036). The sample size for cataract detection on external eye photo was 198 and the P value was 0.102. Error bars represent 95% CI. b)Segmentation dice similarity coefficient. The sample size for segmentation on CFP are: HE (n = 13), SE (n = 14), HM (n = 27) and MA (n = 27). The P value for haemorrhages was 0.083. The sample size was 759 for OCT segmentation. Error bars represent 95% CI. c) Cross-modality disease detection of ciDME that usually need to be diagnosed by OCT with CFP inputs only (left), the sample size was 405 and the P value was <0.001. Cross-modality disease detection of wet-AMD that usually need to be diagnosed by CFP with external eye photo inputs (right) the sample size was 332 and the P value was 0.583. Boxes indicate quartile values and whiskers indicate 1.5×the interquartile range. d) Image-report generation, model performance was evaluated by automatic metrics labelled as the x-axis. The sample size was 500. Boxes indicate quartile values and whiskers indicate 1.5× the interquartile range. e) Head-to-head comparison of answers generated by EyeFM and ophthalmologists, the measurement was summed score for quality, safety and empathy, ranged from 3–15 scores. Presented as Kernel Density Plot. The sample size was 300 for EyeFM and 1200 for ophthalmologists. P values were calculated with two-sided t-test between EyeFM and the better-performed reference model. *** denotes P < 0.001, n.s. represents P > 0.05. CFP, colour fundus photo; OCT, optical coherence tomography; EEP, external eye photo; DR, diabetic retinopathy; DME, diabetic macular oedema; AMD, age-related macular degeneration; MMD, myopic macular degeneration; MA, microaneurysms; HE, hard exudates; HM, haemorrhages; SE, soft exudates.
Extended Data Fig. 5 Workflow for the diagnostic study and management study in the RCT.
Participants included in the trial will first receive diagnosis and report by ophthalmologists by CFP, then receive additional OCT examinations for consultant-level reviewers to assess and revise the diagnosis and report. All diagnosis and reports before revision by consultant-level reviewers will be included in the analysis for correct diagnosis rate, correct referral rate and standardization score of reports. Only participants that are correctly diagnosed as ‘with fundus abnormality’ will be included in the follow-up for patient compliance analysis. CFP, colour fundus photo; OCT, optical coherence tomography.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4, Tables 1–30, protocols and examples.
Source data
Fig. 3, 5 and Extended Data Fig. 4
Statistical Source Data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, Y., Qian, B., Li, T. et al. An eyecare foundation model for clinical assistance: a randomized controlled trial. Nat Med (2025). https://doi.org/10.1038/s41591-025-03900-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41591-025-03900-7