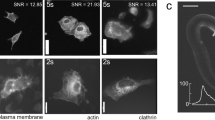
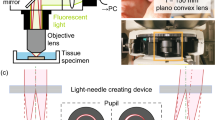
Abstract
Volumetric functional imaging of transient cellular signaling and motion dynamics is often limited by hardware bandwidth and the scarcity of photons under short exposures. To overcome these challenges, we introduce squeezed light field microscopy (SLIM), a computational imaging approach that rapidly captures high-resolution three-dimensional light signals using only a single, low-format camera sensor. SLIM records over 1,000 volumes per second across a 550-µm diameter field of view and 300-µm depth, achieving 3.6-µm lateral and 6-µm axial resolution. Here we demonstrate its utility in blood cell velocimetry within the embryonic zebrafish brain and in freely moving tails undergoing high-frequency swings. Millisecond-scale temporal resolution further enables precise voltage imaging of neural membrane potentials in the leech ganglion and hippocampus of behaving mice. Together, these results establish SLIM as a versatile and robust tool for high-speed volumetric microscopy across diverse biological systems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data underlying the results are publicly available on GitHub at https://github.com/aaronzq/SLIM and via Zenodo at https://doi.org/10.5281/zenodo.15793563 (ref. 70). Source data are provided with this paper.
Code availability
Codes for 3D reconstruction are available on GitHub at https://github.com/aaronzq/SLIM and via Zenodo at https://doi.org/10.5281/zenodo.15793563 (ref. 70) under a BSD-License.
Change history
14 November 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41592-025-02978-8
References
Meng, G. et al. Ultrafast two-photon fluorescence imaging of cerebral blood circulation in the mouse brain in vivo. Proc. Natl Acad. Sci. USA 119, e2117346119 (2022).
Abdelfattah, A. S. et al. Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 365, 699–704 (2019).
Abdelfattah, A. S. et al. Sensitivity optimization of a rhodopsin-based fluorescent voltage indicator. Neuron 111, 1547–1563.e9 (2023).
Hochbaum, D. R. et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods 11, 825–833 (2014).
Wu, J. et al. Kilohertz two-photon fluorescence microscopy imaging of neural activity in vivo. Nat. Methods 17, 287–290 (2020).
Zhang, T. et al. Kilohertz two-photon brain imaging in awake mice. Nat. Methods 16, 1119–1122 (2019).
Wang, Z. et al. Imaging the voltage of neurons distributed across entire brains of larval zebrafish. Preprint at https://doi.org/10.1101/2023.12.15.571964 (2023).
Weber, T. D., Moya, M. V., Kılıç, K., Mertz, J. & Economo, M. N. High-speed multiplane confocal microscopy for voltage imaging in densely labeled neuronal populations. Nat. Neurosci. 26, 1642–1650 (2023).
Sacconi, L. et al. KHz-rate volumetric voltage imaging of the whole Zebrafish heart. Biophys. Rep. 2, 100046 (2022).
Voleti, V. et al. Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0. Nat. Methods 16, 1054–1062 (2019).
Pavani, S. R. P. et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc. Natl Acad. Sci. USA 106, 2995–2999 (2009).
Llull, P. et al. Coded aperture compressive temporal imaging. Opt. Express 21, 10526–10545 (2013).
Wagadarikar, A. A., Pitsianis, N. P., Sun, X. & Brady, D. J. Video rate spectral imaging using a coded aperture snapshot spectral imager. Opt. Express 17, 6368–6388 (2009).
Prevedel, R. et al. Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy. Nat. Methods 11, 727–730 (2014).
Cong, L. et al. Rapid whole brain imaging of neural activity in freely behaving larval zebrafish (Danio rerio). eLife 6, e28158 (2017).
Skocek, O. et al. High-speed volumetric imaging of neuronal activity in freely moving rodents. Nat. Methods 15, 429–432 (2018).
Zhang, Z. et al. Imaging volumetric dynamics at high speed in mouse and zebrafish brain with confocal light field microscopy. Nat. Biotechnol. 39, 74–83 (2021).
Wagner, N. et al. Instantaneous isotropic volumetric imaging of fast biological processes. Nat. Methods 16, 497–500 (2019).
Wang, Z. et al. Real-time volumetric reconstruction of biological dynamics with light-field microscopy and deep learning. Nat. Methods 18, 551–556 (2021).
Lu, Z. et al. Virtual-scanning light-field microscopy for robust snapshot high-resolution volumetric imaging. Nat. Methods 20, 735–746 (2023).
Wagner, N. et al. Deep learning-enhanced light-field imaging with continuous validation. Nat. Methods 18, 557–563 (2021).
Yi, C. et al. Video-rate 3D imaging of living cells using Fourier view-channel-depth light field microscopy. Commun. Biol. 6, 1259 (2023).
Tian, T., Yuan, Y., Mitra, S., Gyongy, I. & Nolan, M. F. Single photon kilohertz frame rate imaging of neural activity. Adv. Sci. 9, 2203018 (2022).
Guo, R. et al. EventLFM: event camera integrated Fourier light field microscopy for ultrafast 3D imaging. Light Sci. Appl. 13, 144 (2024).
Ashok, A. & Neifeld, M. A. Compressive light field imaging. In Proc. Three-Dimensional Imaging, Visualization, and Display 2010 and Display Technologies and Applications for Defense, Security, and Avionics IV Vol. 7690 (eds Bahram, J. et al.) 221–232 (SPIE, 2010).
Babacan, S. D. et al. Compressive light field sensing. IEEE Trans. Image Process. 21, 4746–4757 (2012).
Marwah, K., Wetzstein, G., Bando, Y. & Raskar, R. Compressive light field photography using overcomplete dictionaries and optimized projections. ACM Trans. Graph. 32, 46:1–46:12 (2013).
Antipa, N., Necula, S., Ng, R. & Waller, L. Single-shot diffuser-encoded light field imaging. In 2016 IEEE International Conference on Computational Photography (ICCP) 1–11 (IEEE, 2016).
Liu, F. L., Kuo, G., Antipa, N., Yanny, K. & Waller, L. Fourier diffuserScope: single-shot 3D Fourier light field microscopy with a diffuser. Opt. Express 28, 28969–28986 (2020).
Yanny, K. et al. Miniscope3D: optimized single-shot miniature 3D fluorescence microscopy. Light. Sci. Appl. 9, 171 (2020).
Antipa, N. et al. DiffuserCam: lensless single-exposure 3D imaging. Optica 5, 1–9 (2018).
Feng, X., Ma, Y. & Gao, L. Compact light field photography towards versatile three-dimensional vision. Nat. Commun. 13, 3333 (2022).
Feng, X. & Gao, L. Ultrafast light field tomography for snapshot transient and non-line-of-sight imaging. Nat. Commun. 12, 2179 (2021).
Wang, Z., Hsiai, T. K. & Gao, L. Augmented light field tomography through parallel spectral encoding. Optica 10, 62–65 (2023).
Mandracchia, B. et al. High-speed optical imaging with sCMOS pixel reassignment. Nat. Commun. 15, 4598 (2024).
Guo, C. et al. Fourier light-field microscopy. Opt. Express 27, 25573–25594 (2019).
Llavador, A., Sola-Pikabea, J., Saavedra, G., Javidi, B. & Martínez-Corral, M. Resolution improvements in integral microscopy with Fourier plane recording. Opt. Express 24, 20792–20798 (2016).
Scrofani, G. et al. FIMic: design for ultimate 3D-integral microscopy of in-vivo biological samples. Biomed. Opt. Express 9, 335–346 (2018).
Levoy, M., Ng, R., Adams, A., Footer, M. & Horowitz, M. Light field microscopy. ACM Trans. Graph. 25, 924–934 (2006).
Broxton, M. et al. Wave optics theory and 3-D deconvolution for the light field microscope. Opt. Express 21, 25418–25439 (2013).
Zhou, Y., Zickus, V., Zammit, P., Taylor, J. M. & Harvey, A. R. High-speed extended-volume blood flow measurement using engineered point-spread function. Biomed. Opt. Express 9, 6444–6454 (2018).
Tomina, Y. & Wagenaar, D. A. A double-sided microscope to realize whole-ganglion imaging of membrane potential in the medicinal leech. eLife 6, e29839 (2017).
Briggman, K. L. & Kristan, W. B. Imaging dedicated and multifunctional neural circuits generating distinct behaviors. J. Neurosci. 26, 10925–10933 (2006).
Briggman, K. L., Abarbanel, H. D. I. & Kristan, W. B. Optical imaging of neuronal populations during decision-making. Science 307, 896–901 (2005).
Kannan, M. et al. Dual-polarity voltage imaging of the concurrent dynamics of multiple neuron types. Science 378, eabm8797 (2022).
Buzsáki, G. Theta oscillations in the hippocampus. Neuron 33, 325–340 (2002).
Gu, Z. et al. Hippocampal interneuronal α7 nAChRs modulate theta oscillations in freely moving mice. Cell Rep. 31, 107740 (2020).
Taxidis, J. et al. Voltage imaging reveals hippocampal inhibitory dynamics shaping pyramidal memory-encoding sequences. Nat. Neurosci. 28, 1946–1958 (2025).
Varga, C., Golshani, P. & Soltesz, I. Frequency-invariant temporal ordering of interneuronal discharges during hippocampal oscillations in awake mice. Proc. Natl Acad. Sci. USA 109, E2726–E2734 (2012).
Yoon, Y.-G. et al. Sparse decomposition light-field microscopy for high speed imaging of neuronal activity. Optica 7, 1457–1468 (2020).
Wang, Z. et al. A hybrid of light-field and light-sheet imaging to study myocardial function and intracardiac blood flow during zebrafish development. PLoS Comput. Biol. 17, e1009175 (2021).
Vedula, V. et al. A method to quantify mechanobiologic forces during zebrafish cardiac development using 4-D light sheet imaging and computational modeling. PLoS Comput. Biol. 13, e1005828 (2017).
Zhang, Y. et al. DiLFM: an artifact-suppressed and noise-robust light-field microscopy through dictionary learning. Light. Sci. Appl. 10, 152 (2021).
Pégard, N. C. et al. Compressive light-field microscopy for 3D neural activity recording. Optica 3, 517–524 (2016).
Lin, X., Wu, J., Zheng, G. & Dai, Q. Camera array based light field microscopy. Biomed. Opt. Express 6, 3179–3189 (2015).
Dunsby, C. Optically sectioned imaging by oblique plane microscopy. Opt. Express 16, 20306–20316 (2008).
Zhang, Y. et al. Computational optical sectioning with an incoherent multiscale scattering model for light-field microscopy. Nat. Commun. 12, 6391 (2021).
Zhang, Y. et al. Multi-focus light-field microscopy for high-speed large-volume imaging. PhotoniX 3, 30 (2022).
Li, C., Chen, G., Zhang, Y., Wu, F. & Wang, Q. Advanced fluorescence imaging technology in the near-infrared-II window for biomedical applications. J. Am. Chem. Soc. 142, 14789–14804 (2020).
Miao, Y. et al. Recent progress in fluorescence imaging of the near-infrared II window. Chem. Bio. Chem. 19, 2522–2541 (2018).
Biggs, D. S. C. & Andrews, M. Acceleration of iterative image restoration algorithms. Appl. Opt. 36, 1766–1775 (1997).
Ershov, D. et al. TrackMate 7: integrating state-of-the-art segmentation algorithms into tracking pipelines. Nat. Methods 19, 829–832 (2022).
Tinevez, J.-Y. et al. TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017).
Tomina, Y. & Wagenaar, D. Dual-sided voltage-sensitive dye imaging of leech ganglia. Bio. Protoc. 8, e2751 (2018).
Miller, E. W. et al. Optically monitoring voltage in neurons by photo-induced electron transfer through molecular wires. Proc. Natl Acad. Sci. USA 109, 2114–2119 (2012).
Wetzel, A. W. et al. Registering large volume serial-section electron microscopy image sets for neural circuit reconstruction using FFT signal whitening. In Proc. 2016 IEEE Applied Imagery Pattern Recognition Workshop (AIPR) 1–10 (IEEE, 2016).
Thomson, D. J. Spectrum estimation and harmonic analysis. Proc. IEEE 70, 1055–1096 (1982).
Thomson, D. J. Jackknifed error estimates for spectra, coherences, and transfer functions. In Advances in Spectrum Analysis and Array Processing Vol 1 (ed. Haykin, S. S.) 58–113 (Prentice Hall, 1991).
Nichol, H., Amilhon, B., Manseau, F., Badrinarayanan, S. & Williams, S. Electrophysiological and morphological characterization of Chrna2 cells in the subiculum and CA1 of the hippocampus: an optogenetic investigation. Front. Cell. Neurosci. 12, 32 (2018).
Wang, Z. Source data for manuscript ‘Kilohertz volumetric imaging of in-vivo dynamics using squeezed light field microscopy’. Zenodo https://doi.org/10.5281/zenodo.15793563 (2025).
Acknowledgements
We thank Y. Dong and Y. Zhang at UCLA for their assistance in zebrafish experiments. We acknowledge the David Geffen School of Medicine at UCLA for providing the Zebrafish Core Facilities. This work was supported by the following grants: NIH (grant nos. R01HL165318 (L.G.), RF1NS128488 (L.G.), R35GM128761 (L.G.), R01AI102584 (G.C.L.W.), R01HL129727 (T.K.H.), R01HL159970 (T.K.H.) and T32HL144449 (T.K.H.), R01NS123681 (M.Z.L., P.G.), UM1MH136462 (M.Z.L.), and R34NS128890 (M.K.)). W.C.S. was supported by the National Science Foundation Graduate Research Fellowship Program (grant nos. DGE-1650604 and DGE-2034835) and Ruth L. Kirschstein National Research Service Award ‘Multidisciplinary Training in Microbial Pathogenesis’ (grant no. T32AI007323). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper.
Author information
Authors and Affiliations
Contributions
Z.W. and L.G. conceived of the idea. L.G., D.A.W., R.L., G.C.L.W., T.K.H. and P.G. oversaw the project. Z.W. constructed the microscopes and developed the reconstruction algorithm. R.Z., D.A.W. and Z.W. prepared medicinal leech samples and electrophysiology study. D.E. and L.S. performed mouse surgery. Z.W., R.Z., A.P. and J.W. bred the zebrafish. C.K.L. and W.C.S. prepared bacteria samples. W.K. fabricated the lenslet array. Z.W., R.Z., D.A.W., D.E., L.S., O.B. and E.Z. collected the imaging data. Z.W. and R.Z. processed and analyzed the data. Z.W., R.Z. and L.G. wrote the paper. S.L., M.Z.L., and M.K. generated and tested new reagents. All authors reviewed, edited and consulted on the text.
Corresponding author
Ethics declarations
Competing interests
L.G. has a financial interest in Lift Photonics. However, it was not involved in the research presented in this paper. The other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Robert Prevedel and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Nina Vogt, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 3D voltage imaging of hippocampus pyramidal neurons in awake mice.
a. MIP of SLIM reconstruction of the 3D located neurons. Neuron indices are partially labeled in y-z view to avoid cluttered markers. Representative result from five FOVs using mice labeled for excitatory pyramidal neurons. Scale bar, 100 µm. b. 3D distribution of neuron center locations and their corresponding index. c. Detrended signal traces and the detected spikes labeled in black dots.
Extended Data Fig. 2 Observation of subthreshold oscillations in hippocampus interneurons.
a. Example MIP of identical dataset used in Fig. 4, with neurons of interest labeled with number. Representative result from nine FOVs conducted using mice labeled for OLM interneurons. Scale bar, 100 μm. b. The power spectral densities of 18 neuron traces throughout the entire recording (around three minutes). Red regions denote the frequency band known for theta oscillations. c. Raw traces of ten seconds with red lines plot the band-pass filtered signal in 4–10 Hz.
Extended Data Fig. 3 Comparison between SLIM reconstruction and ground truth.
High-resolution ground truth images are acquired with a reference camera (sharing same objective with SLIM) under either widefield or light-sheet illumination. Sample is axially scanned with a motorized stage to acquire a z stack. SLIM and ground truth image are acquired sequentially on the same sample. a. Tilted mouse kidney slice (FluoCells™ Prepared Slide from ThermoFisher) b. Live mouse hippocampus (CA1, excitatory pyramidal neurons). Green arrows denote representative neuron candidates. Yellow arrows mark the overlaps between sub-aperture views (fabrication tolerance of prisms and holder). When sample exhibits a strong background, this overlap could induce reconstruction artefacts. Blue arrows indicate the MIP artifacts due to the bright boundary of circular FOV. c. Embryonic zebrafish vasculature (endothelial cells, Tg(flk:mCherry)). Scale bar, 100 μm.
Supplementary information
Supplementary Information
Supplementary Figs. 1–18, Notes 1–4 and Tables 1–4.
Supplementary Video 1
3D tracking of blood flow dynamics in the zebrafish vasculature (dorsal view) using SLIM.
Supplementary Video 2
3D tracking of blood flow dynamics in the zebrafish vasculature (ventral view) using SLIM.
Supplementary Video 3
High-speed 3D imaging of a freely moving zebrafish tail using SLIM.
Supplementary Video 4
3D imaging of neuronal action potentials in leech ganglion with SLIM.
Supplementary Video 5
3D imaging of a beating zebrafish heart with SLIM.
Supplementary Video 6
3D imaging of free-swimming Vibrio cholerae bacteria with SLIM.
Source data
Source Data Fig. 3
Source data of Fig. 3.
Source Data Fig. 4
Source data of Fig. 4.
Source Data Fig. 5
Source data of Fig. 5.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Z., Zhao, R., Wagenaar, D.A. et al. Kilohertz volumetric imaging of in vivo dynamics using squeezed light field microscopy. Nat Methods 22, 2194–2204 (2025). https://doi.org/10.1038/s41592-025-02843-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02843-8