Abstract
Cell tracking is an indispensable tool for studying development by time-lapse imaging. However, existing cell trackers cannot assign confidence to predicted tracks, which prohibits fully automated analysis without manual curation. We present a fundamental advance: an algorithm that combines neural networks with statistical physics to determine cell tracks with error probabilities for each step in the track. From these, we can obtain error probabilities for any tracking feature, from cell cycles to lineage trees, that function like P values in data interpretation. Our method, OrganoidTracker 2.0, greatly speeds up tracking analysis by limiting manual curation to rare low-confidence tracking steps. Importantly, it also enables fully automated analysis by retaining only high-confidence track segments, which we demonstrate by analyzing cell cycles and differentiation events at scale for thousands of cells in multiple intestinal organoids. Our approach brings cell dynamics-based organoid screening within reach and enables transparent reporting of cell-tracking results and associated scientific claims.
Similar content being viewed by others
Main
Cell proliferation, differentiation, movement and organization in complex cell lineages are key to understanding organ homeostasis and associated diseases. The development of organoid cultures, which recapitulate key features of organ development ex vivo1,2, has enabled the study of developmental dynamics at the single-cell level using time-lapse microscopy3,4,5,6,7,8. To address the complex challenge of analyzing the dynamics of hundreds of cells in dense three-dimensional (3D) organoid architectures over multiple generations, artificial intelligence-driven semi-automated algorithms have been developed that track cells based on their fluorescently labeled nuclei3,4,9,10,11,12.
However, all current cell-tracking approaches face a fundamental limitation: algorithms output a single tracking solution among many possible solutions, are prone to making errors and yet lack a statistical basis to quantify prediction uncertainty (Fig. 1a). This lack of statistical interpretability makes rigorous analysis based on cell tracks impossible, as the inability to assess the confidence of tracking-based results can lead to unfounded conclusions and, more generally, limits scientific transparency and reproducibility. Finally, the black box nature of cell tracking hampers method development and optimization itself, as it makes it difficult to identify and tackle the true source of tracking errors. By contrast, other widely used bioinformatic methods, such as sequence alignment13,14 or differential gene analysis15, do provide statistics on their output, and the resulting confidence in data interpretation and reporting was crucial to their widespread adoption.
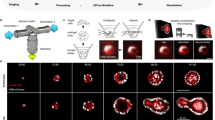
a, Current cell-tracking algorithms convert microscopy images into cell tracks without providing information on accuracy. Yet even single errors can greatly alter the biological interpretation of lineages (here, change in symmetry of divisions). Hence, extensive manual review is required and finally no assessment of statistical confidence can be provided. b, OrganoidTracker 2.0 outputs not only tracks but also associated error rate estimates, greatly aiding data interpretability and transparency. These error estimates also enable drastically reduced manual review or fully automated filtering to achieve high-confidence datasets. c, Method workflow, highlighting two new components (gray boxes): i, Generation of 3D confocal stacks of nuclear marker fluorescence. ii, Neural network detection of nuclear centers. iii, Neural network prediction of cell linking or division probabilities, based on image crops. iv, Constructing a graph representation of the tracking problem, based on predicted link and division probabilities. v, Determination of the globally optimal solution representing the most likely cell trajectories. vi, Estimating link error rates through systematic comparison with alternative tracking solutions. vii, Predicted cell tracks with error rate predictions for individual links.
These problems are particularly acute when studying development and tissue homeostasis, in which an error in even a single tracking step can radically alter biological interpretation (Fig. 1a). For organoids, additional tracking challenges are presented by closely packed nuclei that move rapidly during cell division16. While the recent adoption of neural networks in cell-tracking algorithms has greatly increased tracking quality4,9,10, current methods are far from being free of error, especially in organoids3,4. Existing methods use ad hoc heuristics, such as rapid nuclear volume changes or large cell displacements, to flag potential errors for manual correction3,4. These methods rely on manually set cutoffs and user interpretation, which hampers reproducibility. Moreover, because such heuristics do not provide any measure of confidence in the obtained cell tracks, creating error-free datasets relies on extensive manual curation, up to the point of checking essentially each tracking step. This process can take days for a single 300–500-cell organoid, making tracking applications such as screening different growth conditions or mutant backgrounds prohibitively time consuming.
Here, we present a conceptually new approach: an algorithm that determines both cell trajectories and their error rates (Fig. 1b). Building on our previously developed OrganoidTracker4, we introduced two major innovations: first, we show that neural networks can perform key tracking tasks, such as linking cells between time points and identifying divisions, while providing accurate estimates of the error probability of their prediction. Second, we used concepts from statistical physics, including microstates, partition functions and marginalization, to combine the neural network error predictions into ‘context-aware’ error probabilities that implement our intuition that a low-probability tracking step can in fact be of high confidence, if all alternative cell-linking arrangements are excluded by high-confidence tracks of surrounding cells.
Importantly, these innovations now also enable the reporting of statistical significance. The resulting OrganoidTracker 2.0 can provide error probabilities for any lineage feature of interest, from cell cycles to entire lineage trees. These error probabilities can then be used to assess and report the statistical significance of conclusions based on these tracking features, performing a role similar to that of P values. Our innovations also enhanced tracking performance. First, OrganoidTracker 2.0 is a highly competitive cell tracker, with output tracks containing errors at <0.5% per cell per frame for intestinal organoid data, even before manual curation. Moreover, it drastically sped up this manual curation by focusing it on those parts of cell tracks that had high predicted error rates. A 60-h movie with over 300 cells tracked for over 300 time points was curated in hours rather than days. Second, the resulting method enables fully automated analysis without any human curation by removing, instead of reviewing, the low-confidence parts of cell tracks and using the high-confidence parts for further analysis. Demonstrating the power of this approach, we extracted cell cycle time and rates of differentiation and proliferation for 20 organoids in a fully automated manner, thus opening up the possibility of high-throughput screening of cellular dynamics. OrganoidTracker 2.0 also provides excellent automated tracking for mouse blastocysts and Caenorhabditis elegans embryos, with its performance for the latter ranking as the best-performing tracking algorithm on the Cell Tracking Challenge17. Furthermore, we provide an easy user interface, extensive documentation and straightforward retraining procedures for different biological model systems.
Results
Method overview
Our method is divided into two parts: first, we use neural networks to identify the cells in each frame and predict the probabilities of all possible links between them (Fig. 1c(i–iv)). Next, we use these results to find the most likely tracks and compute their error rates (Fig. 1c(v–vii)). Central to our approach is a probabilistic graph description of the tracking problem18 (Fig. 2a). Here, each node is a cell detected at one time point, while links between nodes represent possible connections between cell detections. To each link, we assign a ‘link energy’, defined as the negative relative log likelihood of a link being true, so that low energy indicates a more plausible link. Similarly, we determine a ‘division energy’ for each node that indicates division likelihood. Expressing predictions of likelihoods as energies allows the use of statistical physics concepts to analyze and combine these predictions. A key innovation is that we employ neural networks to predict these link and division likelihoods based on microscopy data. Here, we leverage a fundamental ability of classification neural networks that use a cross-entropy loss during training, especially when combined with Platt scaling subsequently19,20, namely, that their output scores form accurate probability estimates20, which has thus far not been used in tracking applications. Using an integer flow solver, we find the collection of paths on the graph with the minimal associated energy18, representing the most probable set of cell tracks. Finally, we use the link energies and graph structure to compute context-aware error probabilities for every link in the predicted tracks, thereby providing both cell tracks and their associated error rates. Below, we discuss each step in more detail.
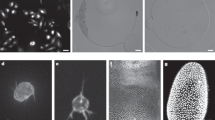
a, Probabilistic graph workflow. Nodes are detected cells, and gray lines are possible links that connect cells between time points. Thicker lines indicate links with lower ‘energy’, that is, more likely. Blue lines represent the globally optimal solution. Cell detection (i) and link and division (div) likelihood (Llink and Ldiv, respectively) prediction (iii) are performed by neural networks. b, Neural network-predicted relative log likelihoods strongly correlate with measured relative log likelihoods (the probability of being true in the manually annotated control) both for links and divisions. Dashed line corresponds to perfect calibration. Data represent n = 5 organoids, with the shaded region denoting standard deviation around the mean. c, A 3D U-Net neural network trained to generate a distance map that indicates proximity to nuclear centers. Cell centers (green squares) are obtained by peak finding. Smaller squares indicate cell centers located below or above the z slice shown. Insets: part of the organoids at higher resolution. Scale bars, 25 µm and 5 µm (inset). d,e, Accuracy of cell detection, compared to OrganoidTracker 1.0, as a function of time (d) and imaging depth (e), for one organoid dataset. Metrics were averaged over ten frames. For e, only cells <40 µm deep were included. f, CNN trained to predict link likelihoods, based on crops centered around two cells detected at subsequent time points. Output images demonstrate a high (green) and low (red) likelihood link prediction, corresponding to true and false links, respectively. Scale bars, 5 µm. g, Link analysis for manually curated data shows that both true (green) and false (red) links are observed for large displacements. Insets: a correct large-displacement link of a cell undergoing division. Scale bars, 5 µm. h, Link prediction accuracy. For all but the smallest displacements, the neural network strongly outperforms predictions based on the ‘smallest-displacement’ criterion, which assigns the link that minimizes displacement as correct. i, Neural network trained to predict division likelihood based on image crops centered at detected cells. Images show three subsequent frames: just before chromosome separation (green border, high predicted likelihood) and before and after (red, low predicted likelihood). Scale bars, 5 µm. j, Fraction of cell crops assigned as dividing (>50% probability) versus time relative to chromosome segregation. Division assignments occur predominantly at the exact measured division time.
Cell detection
We detect cell centers using a 3D U-Net neural network21 (Fig. 2c). Specifically, this network uses 3D images of organoids carrying a fluorescent nuclear marker (H2B-mCherry; Supplementary Fig. 1) to predict a distance map that, for every pixel, records its distance to the closest cell center4,10. Cell centers then correspond to local peaks in this distance map. This approach enables the generation of training data by annotating cell centers, which is less labor intensive than manual 3D segmentation of nuclei. A challenge of distance maps is that cell center peaks for closely packed nuclei blend into one another, causing undersegmentation. We therefore developed an adaptive distance map, in which we assigned increased distance values to pixels that are almost equidistant to two cell centers (Fig. 2c and Supplementary Fig. 2a). This ensured that cells remained well separated in the resulting map, thus reducing segmentation errors (Supplementary Fig. 2b). Overall, the adaptive distance map (together with additional improvements in the training data generation and augmentation pipeline; Methods) substantially improved detection accuracy compared to that of OrganoidTracker 1.0, decreasing the error rate about fourfold. The high accuracy only decreased slightly (99% to 95%) when cell nuclei showed poor signal-to-noise ratio (SNR) after prolonged imaging (>50 h; Fig. 2d) or deep in the imaging volume (>40 μm; Fig. 2e). Finally, predicted cell centroids closely aligned with the center of mass of the 3D nuclear shape of each cell (Supplementary Fig. 3).
Estimating link and division probability
We then construct the linking graph by connecting each node, representing a detected cell, through all potential links, culling links that represented unrealistically large displacements (Methods). Here, links either connect the same cell in two consecutive frames or connect a mother and daughter cell. We designed a neural network that takes in cropped 3D fluorescence images centered on the detected position of each cell for time points t and t + 1 and predicts the likelihood that they represent the same cell (Fig. 2f). The trained network correctly assigned low energy (high likelihood) to links between the same cell, even when the fluorescence signal changed substantially, while assigning high energy (low likelihood) for links connecting a cell to its neighbor, with ‘energy’ defined as the negative relative log likelihood. We compared the network’s performance to the baseline criterion, often used for tracking22, that the links representing the smallest displacement between frames are correct. While links representing smaller displacements (<3 μm) were often true links in ground truth data (Fig. 2g), we also observed true large-displacement links (3–7 μm), which often represented dividing cells and are thus essential for lineage tree reconstruction (Fig. 2g, inset and Supplementary Fig. 4). Only the neural network correctly identified these large-displacement links (Fig. 2h), both for dividing (Supplementary Fig. 4a–d) and nondividing fast-moving (Supplementary Fig. 5a,b) cells.
To determine the likelihood that a node represents a dividing cell, thus connected to its daughters by two outgoing links, we exploited the distinct nuclear morphology of dividing cells with the chromosomal metaphase plate. We designed an additional neural network that used 3D image crops to predict division likelihood, including the previous and subsequent frames to precisely identify the division moment (Fig. 2i). For images at different times relative to division, defined as the last frame before chromosome separation, division assignment (>50% probability) indeed coincided with the moment of division in >90% of cases (Fig. 2j). Moreover, cells at time points before or after division were only rarely assigned as dividing, even when visually similar to cells at the exact division moment.
Prediction accuracy was significantly improved by upsampling challenging cases during training: fast-moving and dividing cells for links (Supplementary Fig. 6) and dying cells for divisions (Supplementary Fig. 7). The ability to tailor training datasets to individual tasks is a major advantage of our modular approach, compared to merging multiple tasks in a single, more complex neural network3,10. Finally, we validated that neural network output indeed represented true probabilities. We binned all possible links based on their predicted likelihood to be correct. For each bin, we calculated the true likelihood, that is, the fraction of links that were correct according to the ground truth data. We found that predicted likelihoods were well calibrated, with predicted and true link likelihood matching for the full likelihood range (Fig. 2b).
Track prediction
To construct cell trajectories, we use a min-cost flow-solver algorithm18 to select the set of links in the probabilistic graph that globally minimize energy and thus maximize the probability of the tracking solution. While close-to-optimal tracks are obtained readily, the algorithm does not guarantee identification of the global optimum, and we typically found minor mistakes, such as link pairs that decrease global energy when swapped. Moreover, flow solvers cannot change the graph structure by adding or merging nodes, causing vulnerability to undersegmentation and oversegmentation, respectively23. We therefore automatically check whether overall probability is increased by swapping link pairs and by adding or merging nodes (Methods and Extended Data Fig. 1a,b). This statistically rigorous and fully automated post-processing procedure substantially increases the duration over which cells can be continuously tracked (Extended Data Fig. 1c).
Context-aware estimation of link error
Central to our approach is estimating the error rate of individual links. The ‘naive’ link likelihoods, as predicted by the neural network, provide information on each link’s error probability but do not take into account the context of the link predictions made for surrounding cells. The importance of context is evident already in manual tracking: here, human trackers typically first establish high-confidence links, which in turn, by reducing the remaining possible links, facilitates subsequent assignment of lower-confidence links. Such contextual information can also be computed in our probabilistic framework. This is illustrated by the simplified graph in Fig. 3a, in which the likelihood of a low-confidence link being true is increased dramatically (from 50% to 98%) in the context of the larger graph, because high-likelihood links exclude all alternative linking arrangements. To generalize this notion (Fig. 3b), we calculate the energy E(Wi) for each possible tracking solution Wi by summing all link and division energies, with the solution likelihood proportional to \({e}^{-E({W}_{i})}\). The ‘context-aware’ likelihood of link A is then given by the total likelihood of all tracking solutions containing that link, \(\sum {e}^{-E({W}_{i,{\rm{A}}})}\), normalized to the sum for all possible solutions, \(\sum {e}^{-E({W}_{i})}\). We call this procedure marginalization, as all other variables are marginalized out to arrive at a single-link error rate estimate without referencing any other links. As computing all possible tracking solutions is unfeasible, we considered only local subgraphs of links less than three steps away (Fig. 3b and the Methods). Increasing subgraphs to four links away did not improve prediction accuracy (Extended Data Fig. 2 and the Methods). For three-link subgraphs, marginalization required <1 h for a 60-h time-lapse dataset.
a, Simplified example explaining marginalization. Lines indicate putative links A–D, with thickness indicating their estimated probability. Link A (red) has a low predicted probability. However, the high-probability link D implies that A is true with high certainty by excluding options containing B and C. The probability P(A|G) that A is true given graph structure G can be calculated by comparing the probability of configurations containing link A and those that do not. b, Schematic outline of marginalization performed on a subset of links around the link of interest. P(A|G) is given by the summed energy of all configurations containing link A normalized to the summed energy of all configurations. c,d, Measured link likelihood versus naive likelihoods predicted by the neural network (c) or context-aware likelihoods calculated by marginalization (d). Data are shown for all possible links (black) or links that are either in the global solution (blue) or not (gray). For naive likelihoods (c), links in the tracking solution are more likely correct than expected, while, for context-aware likelihoods (d), they more closely match measured likelihoods, reflecting integration of graph information. Dashed line represents perfect calibration. Data for n = 5 organoids. Shaded region is the standard deviation around the mean. e, Context-aware likelihoods versus naive likelihoods. Dots are individual links. Lines are averages for true (green) or false (red) links. Marginalization increased the predicted likelihood of correct links while decreasing it for incorrect links. f,g, Number of links versus predicted naive (f) or context-aware (g) link likelihood. In f, while most links in the globally optimal solution (blue) are predicted with high confidence (>99% probability), a fraction have confidence levels similar to those of rejected links (gray). By contrast, for g, virtually all globally optimal links are now predicted with high confidence. h, Fraction of links in the globally optimal solution deemed low confidence (<99% probability). The fraction of low-confidence links that were actual errors compared to ground truth (red) is almost identical to the fraction of errors among all links (triangle), indicating that a <99% probability threshold covers virtually all errors. Marginalization thus reclassified many low-confidence links as high-confidence links but not those that represent errors.
Our marginalization procedure assumes that the individual link and division predictions are independent. However, these predictions are partially based on shared inputs, as the image crops used as input might overlap. Assuming that predictions represent independent evidence causes overconfident error predictions when combined, meaning that links deemed very likely (low negative energy) are more often false and links deemed unlikely (high positive energy) are more often true than predicted (Extended Data Fig. 2). We therefore employed the similarity with statistical physics to introduce a ‘temperature’ T that decreases energies to Ei/T for every neural network prediction i. For T > 1, this reduces the confidence of individual predictions to compensate for the overconfidence introduced during the marginalization procedure. We obtain the optimal value of T by calibrating the marginalized predictions against the ground truth. This employs the same data already used for neural network training and validation without further user input required (Extended Data Fig. 2 and the Methods).
Overall, our marginalization approach borrows conceptually from statistical physics, with each possible tracking solution equivalent to a microstate and the normalization factor to the partition function. From a probabilistic perspective, our method extends the multiplicative opinion-pooling framework24,25, in which different opinions (here, neural network predictions) are combined by multiplying and normalizing their associated probabilities (Methods and Supplementary Discussion).
Evaluation of error rate predictions
We compared both naive and context-aware error rate predictions with measured error rates, obtained by testing their predictions against manually annotated datasets. To avoid bias, these datasets were generated independently from the OrganoidTracker pipeline. We used these ground truth cell centers to generate link and division predictions and calculated context-aware error rates. Naive predicted likelihoods, that is, before marginalization, were already well calibrated, but links identified by the flow solver as part of the globally optimal solution displayed measured likelihoods significantly higher than predicted, while measured likelihoods were lower than predicted for links rejected from the global solution (Fig. 3c). This matches our intuition that the graph contains additional information on link likelihood (Fig. 3a), as the flow solver selects links based on complete graph information, while the neural network uses only on local image information. By contrast, context-aware link predictions had strongly improved confidence, reducing the mismatch between predicted and measured likelihood for both flow-solver-selected and -rejected links (Fig. 3d). Moreover, incorporating graph context specifically increased the predicted likelihood of true links while decreasing it for false links (Fig. 3e).
The improved context-aware link predictions have substantial practical advantages for error correction. It strongly increased the differences in likelihood between links rejected or selected from the global solution (Fig. 3f,g). For naive predictions, a large fraction (6%) of links selected by the flow solver must be reviewed when using <99% predicted probability as the threshold for manual curation. For context-aware predictions, this reduced substantially (1%), while practically all true linking mistakes (0.12%) were still detected (Fig. 3h). Many more links (~25%) must be reviewed to achieve similar accuracy using a cell displacement-based heuristic (Supplementary Fig. 8). Our marginalization procedure specifically benefitted challenging links representing large cell displacements (Supplementary Fig. 5c). As a final control, we tested the marginalization procedure in the context of our full pipeline by creating new ground truth datasets for three organoids through manual curation, resulting again in well-calibrated error rates (Extended Data Fig. 3). Lastly, we reduced computation time without reducing accuracy by excluding highly unlikely links before marginalization, as this did not significantly impact error rate computation (Extended Data Fig. 3 and the Methods).
High-level error probabilities for lineage features and manual curation
We ran our pipeline on a ~60-h time-lapse dataset of a representative organoid (Supplementary Videos 1–3). The rate of predicted potential errors (defined as <99% probability links) was 1.5% per cell per frame after removing tracks deep in the imaging volume, with potential errors predominantly, but not exclusively, concerning divisions (Fig. 4a). These error rates can be propagated to complex downstream lineage features as \(p=1-\prod _{i}{P}_{i}\), where Pi are the context-aware probabilities of all links i of the lineage feature of interest and p is the probability that the feature is not correctly tracked (Extended Data Fig. 4). These high-level error probabilities enable users to assess the statistical significance of, for example, individual cell cycles, the observation that cells are sisters or even entire lineage trees (Fig. 4a,b and the Methods). These probabilities can thus function similarly to P values, although we note that they do not follow from a hypothesis-testing framework. This approach also enabled the identification of high-confidence lineage fragments (p < 0.01, calculated over all links within the fragment), yielding stretches containing multiple cell cycles from uncurated tracks (Fig. 4b and Supplementary Fig. 9).
a, Selected lineages before review, with color indicating associated error rates. **, Flagged error with a clearly erroneous lineage structure (unrealistically short cell cycle); *, Not identifiable as an error based on lineage structure alone. Error probabilities (perr) are calculated for entire lineage trees by combining underlying error rates. b, Blue lineage fragments are high confidence (<0.01 error rate). Users can identify high-confidence cell cycles (black arrow) or sister pairs (gray arrow) without manual review. c, Lineage trees after manual review. Gray lineage sections were added following curation. Compared to a, error probabilities now indicate high confidence in the lineages. d, Characterization of potential errors. Links flagged as potential errors either represent (dis)appearing cells (blue) or are low confidence (red). A substantial proportion of potential errors represented short tracks of cellular debris (gray), with no impact on lineage trees when removed and only few actual errors that required correction. e, Three-dimensional reconstruction with colors indicating cells in the same lineage. f, Automated analysis without manual review by filtering out low-confidence links and performing survival analysis on the resulting, partly censored data. g,h, Survival curve of the fraction of cells not divided at time t after birth (g) or after the sister’s division (h). Shown are manually annotated (gray) and automatically filtered (red) data for a single organoid. Vertical dashed line denotes average cell cycle duration, while the horizontal line shows the inferred fraction of cells that stop dividing. Proliferation ceases in 32% of cells (g), while 97% of sister cells divide within a 10-h window of one another (h), highlighting the dominance of symmetric divisions in intestinal organoid growth. Shaded region, 95% confidence interval of the surviving fraction estimate. i, Lineage dynamics parameters obtained by fully automated (red) or manual analysis (gray) show excellent agreement. j,k, Automatically obtained cell cycle duration and its difference between sisters (j, n = 20 organoids) and the fractions of cells that cease proliferation and of asymmetric sisters in which only one cell divides (k, n = 19 organoids). Dots represent individual organoids, and error bars are the standard deviation around the mean.
Our accurate knowledge of link probabilities implied that focusing manual correction only on low-confidence links should suffice to obtain error-free tracks. To test this, we manually reviewed all links with <99% probability. We also reviewed all beginnings and endings of cell tracks mid-experiment (0.9% per cell per frame), which represented cells dying, entering or exiting the imaging volume or cell detection errors. Only a fraction represented true linking or detection errors (0.3% per cell per frame; Fig. 4d). However, correcting the few true errors strongly improved the lineage trees, complementing them with previously unconnected subtrees (Fig. 4c and Supplementary Fig. 10), underscoring the importance of identifying even infrequent tracking errors. Finally, independent manual tracking of cells in the lineages of Fig. 4c yielded identical trees. Correction required ~4 h for a dataset in which ~300 distinct cells were tracked in a ~60-h time window (Fig. 4e, Supplementary Fig. 10 and Supplementary Video 4). When we calculated error probabilities for cell lineages after manual correction, assigning a probability of one to manually corrected links and recomputing the marginalized link likelihoods (Methods), we found low values (p < 0.05) for all analyzed lineage trees (Fig. 4c), indicating high resulting confidence.
Fully automated lineage tracking by error filtering
Manual curation is typically a prerequisite for analysis of 3D cell-tracking data. Our ability to accurately estimate linking error rates enables a new and fundamentally different approach: to remove low-confidence track fragments and analyze only the remaining high-confidence fragments (Fig. 4b,f). For fragments that are high confidence from division to division (Fig. 4b), properties such as cell cycle durations could be directly measured and compared between different organoids. We employed survival analysis (Methods), a statistical framework for dealing with censored (incomplete) data26,27,28, to quantify a broad range of lineage properties while also incorporating information from lineage fragments containing incomplete cell cycles. Specifically, we generated Kaplan–Meier survival curves to estimate the fraction of nondivided cells as a function of time since cell birth (Fig. 4g), using all high-confidence track fragments that included at least one birth. This survival curve plateaued at 32%, representing the fraction of cells that do not divide again and, hence, have differentiated. We can compute cell cycle duration from the survival curve’s decrease in time (Methods), yielding a duration of 17 ± 2.8 h. When we extended this analysis to sisters, using sister pair fragments to generate survival curves relative to the time of sister cell division (Fig. 4h), the curve plateaued at 3%, representing the small fraction of sisters in which one proliferated and the other did not. This high symmetry between sisters is consistent with recent work5,6. Moreover, the survival curve’s steep decrease indicated highly similar cell cycle duration between sisters, with <2.5 h between sister cell divisions. Overall, survival curves generated from automatically filtered and manually tracked data showed an almost exact overlap (Fig. 4h). Finally, we demonstrated the automated nature of this approach by analyzing 20 different organoids (Fig. 4j,k). We consistently found similar parameter values and survival curves, even as organoids displayed differences in size and morphology, indicating that the underlying lineage dynamics is independent of this morphological variation (Fig. 4j,k and Extended Data Fig. 5).
Out-of-sample capabilities
We tested the performance of our neural networks on out-of-sample data, which can degrade cell detection and linking performance, leading to poor tracking, or yield inaccurate error probability predictions. We first examined the influence of biological variation by exposing organoids to the cell cycle inhibitor palbociclib. Palbociclib exposure changed cell appearance and dynamics, with cell division inhibition causing smaller nuclei and reduced movement. Nonetheless, cells were readily tracked through ~40-h lineages without manual curation (Extended Data Fig. 6a), with automated lineage analysis by error filtering demonstrating the expected cell division inhibition (Extended Data Fig. 6b,c). Manual curation of part of the data revealed that error rates remained well calibrated (Extended Data Fig. 6d).
We next tested the impact of using a different confocal microscope (Methods). Differences included lower pixel resolution, lower SNR and an objective with higher working distance, with the latter enabling imaging cells deeper (60 μm, about ten cell diameters) in the organoid. After background subtraction and spatial rescaling to match the training data image resolution (Methods), cells could be tracked through ~40-h complex lineages without curation (Extended Data Fig. 7a,b), even at a depth of 50 μm where SNR was low (Supplementary Videos 5 and 6). Here, manual curation revealed slightly overconfident error predictions (Extended Data Fig. 7c). However, we recovered perfect calibration simply by recalibrating the scaling temperature T used during marginalization, without any neural network retraining. This recalibrated scaling temperature was obtained by manually reviewing ~200 links (<2 h of work; Methods) and did not differ between different organoids and time points (Extended Data Fig. 7d–f). Finally, we note that, even without recalibration, deviations in predicted error rates were small, with only 0.06% of links erroneously not flagged for manual curation (Extended Data Fig. 7c). Indeed, automated lineage analysis by error filtering gave almost identical results before and after recalibration (Extended Data Fig. 7g,h), suggesting that, while recalibration is generally desirable, the impact of bypassing this step is limited.
Finally, we examined performance on a non-organoid model system, using published light-sheet microscopy data of mouse blastocysts29. Using the above approach, we could track most cells in individual blastocysts through ~25-h lineages, corresponding to the 16–64-cell stage, and with low error rates (Extended Data Fig. 8a,b and Supplementary Videos 7 and 8). Blastocyst cells moved more rapidly than intestinal organoid cells, with displacements often larger than the typical nucleus diameter, but were still linked correctly (Methods and Extended Data Fig. 8b–d). Manual correction revealed minor deviations from perfect calibration for error predictions that were corrected by recalibrating the scaling temperature (Extended Data Fig. 8e,f).
The versatility of our algorithm without neural network retraining contrasts with the typical workflow for machine learning-driven 3D cell tracking, in which, for out-of-sample data, new neural networks are (re)trained3,9,30.
Neural network retraining
We examined the performance of our full pipeline on an imaging dataset that required retraining of the underlying neural networks, focusing on a confocal time-lapse microscopy dataset of C. elegans embryogenesis hosted by the Cell Tracking Challenge17,31. We trained cell detection, division and link prediction neural networks with only minimal changes to the training procedure (Methods). We found that our method here performed as well as for intestinal organoid data, generating cell tracks spanning up to seven generations (Extended Data Fig. 9 and Supplementary Video 9), even though training data were limited in comparison. Upon manual review of all <99% confidence links and cell (dis)appearances, corresponding to 0.9% of total links, the resulting data exactly reproduced the known C. elegans lineage structure, while predicted error rates were well calibrated. Independent verification of our automated tracking results before any correction by the Cell Tracking Challenge confirmed the quality of our predictions, ranking us first in tracking performance.
Discussion
In this study, we presented a conceptual innovation in cell tracking: whereas existing algorithms typically generate tracks with minimal information on correctness, OrganoidTracker 2.0 instead estimates the confidence in its predictions. Our approach exploits neural networks to predict linking and division probabilities based on 3D microscopy data and uses statistical physics concepts to adjust these probability estimates based on information of surrounding cells. This enables highly efficient manual curation, by only correcting a minority of low-confidence tracking steps, or fully automated analysis, by using only high-confidence track fragments. It also enables computing error probabilities for any tracking feature, which function akin to P values, allowing researchers to report the statistical significance of their cell-tracking results and associated scientific claims, which we believe will be important in further stimulating the adoption of cell-tracking methods in biology. Our approach is readily extended to cell tracking in other contexts, such as two-dimensional cultures or embryos. OrganoidTracker 2.0 is freely available, with extensive documentation and a user-friendly graphical user interface (GUI)4.
Predicting well-calibrated error probabilities required using distinct neural networks for different tasks rather than a single neural network for cell detection and linking simultaneously3,10. This modularity brings further advantages. First, this enabled task-specific optimization both of network architecture and training data, for instance, by upsampling the number of challenging division events when training the division network. This optimization is greatly aided by the fact that these subtasks have easily interpretable probabilities as their output, which allow their isolated evaluation. Second, each network can be swapped with other implementations30,32,33,34,35,36 tailored to different model systems, as long as they provide well-calibrated probabilities. Finally, it allows extending our approach with additional neural networks to predict probabilities of other events that impact cell tracking, such as cell death, cell extrusion or abnormal divisions37.
Our ability to predict error probabilities represents a fundamental advance in the cell-tracking field. Current state-of-the-art 3D cell tracking typically relies on heuristic rules to identify tracking errors, such as flagging unrealistically large displacements or short cell cycle times3,4, although more systematic track quality measures were developed for 3D particle tracking38. Recent 3D cell-tracking algorithms used neural networks for cell linking35 and detection30 that provided approximate information on link and division probability but not in a manner that supports calculating error rates and statistical significance. For two-dimensional cell tracking, studies used approaches such as linear regression, Bayesian analysis, random forests or Kalman filters32,33,34,39,40,41 to predict link and division likelihoods, sometimes even explicitly calibrating these outputs33, but did not provide error rates or otherwise quantify statistical significance based on these. The key enabling step here is our marginalization procedure (Fig. 3), which increases prediction confidence by incorporating the contextual information provided by linking information of surrounding cells. Without marginalization, too many links erroneously ranked as low confidence for the error probabilities to be useful in subsequent analysis (Fig. 3). Our marginalization procedure is independent of how link probabilities are calculated and hence could benefit other (cell-)tracking algorithms.
Addressing inevitable cell-tracking errors typically requires labor-intensive manual review3. Our error rate predictions strongly reduced manual curation time by focusing exclusively on uncertain links, with a 60-h time-lapse movie of intestinal organoids with ~300 cells requiring only 4 h of manual review instead of days (Fig. 4). Alternatively, selecting only high-confidence fragments of cell tracks or lineages allowed the extraction of lineage features and relationships without human curation. Using this approach, we extracted key features of cell proliferation control, such as cell cycle length, cell cycle arrest rate and cell cycle correlations between sister cells at high throughput (thousands of cells across 20 organoids, ~1 h of computation time per organoid on a desktop computer). This automated analysis could be extended to other biological events, such as cell death or cell cycle stages, when combined with fluorescent markers28,42 or neural networks that can detect these events37. Moreover, it enables systematic characterization of cell proliferation parameters or other features under different conditions43, such as the addition of signaling inhibitors or drugs. These experiments seem especially promising in cancer research, in which studies have demonstrated the power of microscopy-based screens of cancer organoid shape and size44, but for which single-cell analysis at scale is not yet feasible45,46.
Our methods functions over a range of systems and image modalities, provided the nuclear signal quality is similar to what is used for standard manual annotation. Integration of artificial intelligence-driven image restoration, which allows denoising and deblurring47, or 3D ‘cell painting’, which reconstructs nucleus positions based on transmitted light images48, could push beyond this limit. Our algorithm processes data on the basis of the full imaging volume, which renders the analysis of very large volumes (gigabytes of data per frame) prohibitively memory consuming. This might be addressed by combining our framework with approaches that tile data into manageable subvolumes10. Further improvements could come from replacing our convolutional neural networks (CNNs) with transformer-based architectures, which can integrate more complete temporal information in their cell-tracking predictions49,50, incorporating, for example, information on long-term tissue flow. Finally, to calculate error probabilities, we implemented the required marginalization step simply by considering all potential tracking solutions in a local neighborhood, which is computationally intensive and limits the degree of context that is integrated in the error prediction. We speculate that the analogies with statistical physics can be exploited to establish algorithms that sample the space of possible tracking solutions more efficiently, similar, for example, to the Metropolis–Hastings algorithm51,52.
Our results raise fundamental issues regarding the reporting of cell-tracking-based results. For small datasets, manual curation may be performed at least on a limited number of key features such as divisions. However, for larger datasets, such as embryo or gastruloid systems53 or screens involving many conditions, this approach is no longer feasible. Yet, once established, reported tracking results are often treated as a given, without insight into the uncertainties. Currently, confidence of these results and associated claims can only be assessed by studying the original microscopy images, which is typically infeasible. The ability to calculate error probabilities, as we advance here, will be of general importance to mitigate this issue. Similar to any other form of quantification in science, such error probabilities or error probability cutoffs should be reported for displayed cell tracks and lineage trees and for lineage features, such as cell cycles. Reporting error probabilities of published tracking data will also be crucial for data sharing by enabling external users to assess confidence in different features of the data, even without access to the underlying microscopy images. Our work here now provides the conceptual framework and computational tools to extend this approach to a broad range of cell-tracking applications.
Methods
Organoid culture
Mouse intestinal organoids with an H2B-mCherry reporter were used, gifted by N. Sachs and J. Beumer (group of H. Clevers, Hubrecht Institute). Organoids were grown embedded in membrane extract (BME, Trevigen) in medium consisting of murine recombinant epidermal growth factor (50 ng ml−1, Life Technologies), murine recombinant Noggin (100 ng ml−1, PeproTech), human recombinant R-spondin 1 (500 ng ml−1, PeproTech), N-acetylcysteine (1 mM, Sigma-Aldrich), N2 supplement (1×, Life Technologies) and B27 supplement (1×, Life Technologies), GlutaMAX (2 mM, Life Technologies), HEPES (10 mM, Life Technologies) and penicillin–streptomycin (100 U ml−1, 100 μg ml−1, Life Technologies) in Advanced DMEM/F-12 medium (Life Technologies). Organoids were kept in incubators at 37 °C with 5% CO2. The medium was changed every 2 d. Each week, organoids were mechanically broken, and the fragments were reseeded.
Sample preparation
Organoids were seeded around 2 d before imaging in four-well chambered coverglass (#1.5 high-performance coverglass) from Cellvis. For the organoids to move within the lens working distance and minimize the required laser power, we placed the sample on a cold block (~4 °C) for 10 min after seeding. In this manner, the organoid fragments could sink to the bottom before the gel solidified. Afterward, the BME gel was allowed to solidify at 37 °C for 20 min before adding medium.
Microscopy
Imaging was performed on a Nikon A1R MP microscope with a ×40 oil-immersion objective (numerical aperture, 1.30). Around 30 z slices with a step size of 2 µm were taken per organoid every 12 min, with a pixel size of 0.32 µm2. For the low signal-to-noise data, imaging was performed with a Leica TCS SP8 microscope with a ×40 water-immersion objective (numerical aperture, 1.10) with a pixel size of 0.4 µm2.
Computational resources
All analysis described was carried out on a desktop computer with a dedicated graphics card (Nvidia RTX 2080 Ti).
Intestinal organoid training data
Our training data consisted of nine different tracked crypts together with nearby villus regions. Time-lapses were between 16 h and 65 h long, with the full dataset totaling 281 h (1,405 frames). For a given frame, around 150 cells were annotated, meaning that on the order of 200,000 cell detections and links between are present in the training data. This is the same dataset used to train the original OrganoidTracker4; therefore, we can confidently say that any improvements are due to the new algorithm and not because of an expanded training dataset. All training data were generated in the context of an earlier publication5.
Statistics and reproducibility
Representative images (Fig. 2f,g,i) were chosen from hundreds (for divisions) or tens of thousands (for links) of similar-looking images. Random lineages (Fig. 4a–c) were randomly selected from lineages that contained at least one cell at the end point, the ancestry of which could be tracked completely in the manually corrected data.
General neural network training and prediction procedure
The input during both training and predicting for all neural networks consists of a list in which each item references an image frame together with any data needed to create the final neural network input (that is, a list of cell centers around which to crop). Only during training and prediction are image frames loaded, and the input data are generated to minimize the memory footprint. All data augmentation during training is performed at runtime for the same reason. Image frames can be loaded from .tiff files but also from common platform-specific file formats like .lif (Leica) or .nd2 (Nikon) to avoid the need for data conversion.
Before training the neural network, the input list is randomized and split into training and validation sets (80% versus 20%). After training with the link and division detection data, we perform a simple Platt scaling based on the validation dataset to ensure that our predictions are well calibrated19. During Platt scaling, we try to maximize the likelihood of the ground truth data (x) given our scaled predictions (p*):
with, for a given link or division prediction i:
where the scaled predictions are given in terms of the original predictions, p, by (with A and B to be optimized):
The maximum likelihood is then found by minimizing the cross-entropy loss between x and p*:
Gradient descent is performed using the Adam optimizer for all neural networks. The full network architectures can be found on our GitHub (https://github.com/jvzonlab/OrganoidTracker).
Cell center detection: generating training data
To detect cell centers, we use both the frame at the time point of interest and the subsequent frame to give the neural network access to dynamic information. We crop the images to a box that contains all annotated cell centers to avoid learning on unannotated regions. Images are then normalized, after which random crops (32 × 96 × 96 × 2t) are made. Users can set arbitrary time windows and crop sizes when training their own neural networks.
To augment the data, these crops are randomly flipped along the x or y axis (50% of cases) or randomly rotated and scaled (by a random factor between 0.8 and 1.2). Further augmentation is performed by randomly changing the contrast by exponentiation of the intensity values by a random number (between 0.8 and 1.2). The fluorescence intensity decay with increasing image depth can vary greatly between imaging settings. We therefore also augment the data by increasing the decay in intensity with depth by a random factor, such that the deepest frame can have up to a fourfold reduction in intensity.
Cell center detection: distance map and weights
The neural network is trained to predict the distance to the nearest cell center for every pixel in the image. The distances are transformed by a Gaussian function to give rise to diffuse spots centered around cell centers. This approach has achieved success in many cell localization algorithms when the full segmentation of cells is not available4,10,54. We improve this approach by also taking into account distances to nearby cells other than the closest one. By increasing the distances (and thereby decreasing intensities in the distance map) for pixels that are close to another cell, we ensure that the Gaussian spots remain well separated. The mathematical description of the ‘adaptive’ distance d is given by:
in which dmax represents the maximum radius within cell centers still relevant in computing the distance value for a pixel. It can be chosen up to the minimum distance between two cell centers before spots will overlap. The first term measures the distance to the closest cell center, while the second term increases this value if other cell centers are also within dmax.
The intensity values in the distance map are then given by:
in which we choose r to be \(\sqrt{1/8}\) dmax to produce well-separated spots.
The calculation of the distance map is carried out at runtime on the GPU for maximum efficiency. It can be implemented using only convolutional operations by replacing the minimum operator in the equation above with a pseudominimum (soft-min function).
Our algorithm allows users to only partially annotate datasets, reflecting the fact that most existing manually tracked data are often focused on a limited region of interest due to time considerations. Training on partial annotations was enabled by assigning large weights to pixels in the annotated regions versus the background during training. To assign these weights, we change our distance map so that pixels with multiple cells nearby have lower distance values associated with them:
We then use these distances to calculate the weight values:
where b is a small weight assigned to background pixels. By giving some weight to the background, the neural network can learn to ignore debris and imaging artifacts outside the foreground. For the intestinal organoid data, b is chosen such that half of the total summed weights is associated with annotated nuclei and half with the much larger background region.
Cell center detection: neural network
The neural network used for cell detection is very similar to the 3D U-Net used in the previous OrganoidTracker4. The different time points in the input are treated as different channels. A new element in the network is a final smoothing layer (convolution with a Gaussian kernel with a pixel width of 1.5). Because the center point annotation is inherently noisy (not pixel perfect), the predicted output should be smooth. By enforcing this explicitly, we reduce overfitting and speed up the training.
Cell center detection: peak finding
From the predicted distance map, we localize the cell centers by using a peak-finding algorithm, as described before4. Peaks within a certain radius (half the typical distance between nuclei) of other higher peaks are excluded by the peak-finding process to avoid oversegmentation due to noise in the predicted distance map.
During cell division, cells round up and their distance to other nuclei increases. At the same time, cells are more prone to oversegmentation as H2B fluorescence is not uniformly distributed anymore because of chromosome condensation. To counteract this, we revisit the cell detections after we have predicted the division probabilities (see below) and merge dividing cell detections (defined as having a division probability greater than 50%) that are closer than 5 µm from each other.
Cell center detection: evaluation
Cell center detection was evaluated as previously described4. We compared predicted data with partially annotated manual datasets. The evaluation data consisted of five different organoids, imaged on different days, for which at least one crypt was fully tracked. The organoids were tracked for between 90 and 320 frames.
For every cell center in the manually annotated dataset, we check whether there is a predicted cell center within 5 µm; these count as true positives. A predicted center can only match a single-cell center in the manual data. Unmatched manual annotations are false negatives. Predicted cell centers that remain unmatched and are within the manually annotated region (distance of 5 µm from an annotation) are counted as false positives. Consigning the evaluation to annotated regions means that mistakes far from the epithelial layer are ignored (that is, debris recognized as a nucleus), but these are both rare and generally irrelevant for tracking.
Recall is calculated by dividing true positives by the total number of manual annotations. Precision is defined by dividing the false negatives by the amount of predicted cell centers within the annotated region. Accuracy is the number of mistakes over the sum of all observations (true positives, false positives, false negatives).
To test the effect of our ‘adaptive’ distance map, we also trained a network on a target mapping that consisted simply of Gaussian spots around the cell centers. For these spots to not overlap, we had to half their radius relative to the ‘adaptive’ version. The pixel weights were kept the same (Supplementary Fig. 2).
Cell center detection: Cellpose comparison
We used the Cellpose 3D module36 to produce nuclear masks for three time frames of our test dataset. The Cellpose algorithm was run from a dedicated Cellpose plugin in the OrganoidTracker GUI. We used an expected nucleus diameter of 25 pixels. After obtaining nuclear masks, we computed the centroid positions as the center of mass of each 3D mask. We manually removed Cellpose centroids that correspond to oversegmentation or undersegmentation. These validated centroids were then compared to the OrganoidTracker predictions. The analysis was limited to a tissue depth of 15 µm, avoiding the poor Cellpose segmentation for higher depths.
Link detection: proposing possible links
To avoid examining extremely implausible links, we propose links based on the distance between the subsequent cell detections. During both training and prediction, we only consider links from a cell detection to a cell detection in the next frame that are at most two times farther away in distance than the closest cell in the next frame.
Link detection: generating training data
The input of the neural network for link prediction consists of a crop centered around a cell center, a crop around the cell detection in the subsequent frame and a vector describing the distance in pixels between the cells. The two crops are 16 × 64 × 64 in size and both contain the two time points containing the cell center detections. Users can set arbitrary time windows and crop sizes when training their own neural networks.
Data are augmented in the same way as during cell center detection, except that we do not vary the decay in intensity with depth, as the crops are much smaller in the z dimension. Instead, we increase the range in which we vary contrast (exponentiation by a number between 0.5 and 1.5).
To aid prediction, we provide the neural network with direct information about the direction of movement by adding the displacement vector to the neural network inputs beside the crops around the cell centers. It is known that CNNs have trouble integrating information in the form of Cartesian coordinates55. We therefore add an extra three channels to both crops. These contain, for each pixel, the x, y and z distances, respectively, to the other cell center detection in the proposed link.
We upsample difficult cases, cells that are dividing (within a window of an hour around cell division) or move a considerable distance (more than 3 µm, less than 7 µm) by replicating these five times in our training data.
Link detection: neural network
The first part of the neural network for link detection consists of two CNNs. To maximize the amount of information extracted, one CNN takes in the concatenated crops while the second CNN takes as input a single crop (two identical copies of the second CNN are available to analyze both crops). This means that one CNN can integrate pixel information between crops and directly assess how similar the two cell detections at subsequent time points are. The other CNN is forced to focus on a single crop, which could, in combination with information about the direction of movement, already be enough to assess the link probability.
The features extracted by the CNNs in combination with the displacement vector are then fed into multiple densely connected neural network layers to yield a prediction.
Link detection: evaluation
To evaluate the link neural network, we used the same set of evaluation data as used in evaluating the cell center detection. See the main text for the evaluation procedure.
To test the effect of adapting the training data, we also trained a link detection neural network without upsampling difficult cases (Link detection: generating training data). We then compared accuracy, precision and recall across all evaluation organoids (Supplementary Fig. 6).
Division detection: generating training data
The input of the division detection neural network is a crop (12 × 64 × 64) centered around a cell center, with the previous and subsequent frames included for dynamic information. Data augmentation is carried out in the same manner as during link detection training.
To avoid a too low frequency of images related to cell division, we upsample cells in the process of division (within a 1-h window around the nucleus dividing) by replicating them ten times in our training data. We also upsample all dying cells (cells with tracks ending before the end of the experiment), as these can closely resemble dividing cells. From all other cell detections, which are often trivial to predict as nondividing, only a random subset is included so that they make up 20% of the total dataset.
Division detection: neural network
The design of the division detection neural network mimics that of the link detection network. A CNN extracts features that are then fed into a dense layer to generate the prediction. The main difference is that, due to the limited nature of the division datasets (there are only hundreds of divisions present in our training data), we employ only a single dense layer to avoid overfitting.
Division detection: evaluation
To evaluate the division neural network, we again used the same set of evaluation data used in evaluating the other neural networks. See the main text for the evaluation procedure.
To test the effect of adapting the training data, we also trained a division detection neural network without upsampling difficult nondividing cases (‘Division detection: generating training data’). We replaced these difficult cases by randomly selected cell centers so that divisions make up the same fraction of the training data as in our normal training procedure. If we would truly train on an unbiased sampling of the data, so that nondivisions make up the vast majority, this would cause the training procedure to not converge. We then compared accuracy, precision and recall across all evaluation organoids (Supplementary Fig. 7).
Graph description
In our graph description of the dataset, we follow the framework developed ref. 18. Here the nodes of the graph are the detected cell centers and the edges are the proposed links. These edges have an associated energy penalty that is the relative negative log likelihood that the link is true as predicted by the neural network. The nodes have an associated division penalty, which is again the negative relative predicted log likelihood.
Within this framework, we also have to assign energy penalties to the events in which a track disappears or appears or when a cell detection is a false positive. A track can disappear when a cell dies or its next position is not detected. The disappearance probability is thus the combination of the death rate and the false negative rate of the neural network. Here, the latter makes the dominant contribution. Tracks can appear when their previous position is not detected, which again relates to the false negative rate. The probability of a cell detection being spurious is given by the false positive rates. All these rates can be estimated from the validation of the cell detection neural network and are around 1%. Varying these probabilities within an order of magnitude (3% to 0.3%) does not significantly affect track prediction or the marginalization procedure (not shown).
To account for cells appearing or disappearing because they are close to the edge of the imaging volume and can leave the imaging volume, we assign lower (dis)appearance penalties (corresponding to a 10% chance of (dis)appearance) to cell detections at the edges of the volume.
One could imagine a neural network that would assign explicit probabilities to the correctness of cell detections so that we could use node-specific (dis)appearance penalties. This should lead to minor improvements in track quality, but such an approach would have several drawbacks. First of all, training data are limited because the cell detection network makes few mistakes. Furthermore, such a neural network would have to be retrained every time a new cell detection network is trained, as it is specific to the type of mistakes that that network makes. Integrating a neural network to identify dying cells and adapt the disappearance probabilities accordingly would be more feasible37 but of limited use due to the rare nature of cell death in our system.
In principle, the predictions made by division and link detection neural networks are probabilities conditional on the correctness of the underlying cell detections, because only correct cell detections are in the training data. It is possible to assign energy penalties in such a way that they represent probabilities of a link or division conditional on the existence of the node it is coming from by combining the chance that a link is incorrect and that its source node does not exist in a single energy penalty. This could avoid including some oversegmentations that persist over multiple subsequent frames and have high-probability links between them in the tracking solution. But including correct links between oversegmented cells is in our case actually the preferred behavior. Not including these links would hamper our approach of solving these oversegmentations during post-processing (see below). This does mean that, after marginalization, we also have to interpret the predicted error rates as the chance that the two different cells associated with the detections are not linked, not the chance that the link is ‘incorrect’ because one of the two detections is due to an oversegmentation. Because oversegmentation on its own already introduces errors by definition, as a track caused by oversegmentation both has to appear out of nowhere and disappear again, this will not cause any missed errors.
Flow solver
We use the flow solver developed in ref. 18 to find the most likely set of tracks. To help it converge to an optimal solution, we prune the graph of high-energy edges. We do this by comparing every edge to its alternatives: links having the same source or target nodes. If a link with a much lower penalty is available (>4.0 difference, corresponding to a 10,000 times more likely link), we remove the edge. This was not done during the marginalization evaluation (Fig. 3), in which link removal such as this would introduce a bias in the nonmarginalized probabilities for very unlikely links. Potential divisions that have a probability below 0.01 are also removed.
The flow solver sometimes has trouble converging or halts prematurely especially in the presence of a large number of low-certainty predictions. To circumvent this, users can also use the Viterbi-style algorithm proposed by Magnusson et al.41 as implemented by Haubold et al.
Fine-tuning flow-solver solution
Because the flow solver does not guarantee an optimal solution, we fine-tune our solution by checking, for every link, whether removing it and replacing it with an appearance and a disappearance would lower the total energy. We then also look at pairs of links in the solution that connect two nodes at time point t with two nodes at time point t + 1 and check whether they should be replaced with a pair of edges that connect the nodes the other way around. We perform three cycles of this pruning and swapping of links.
Solving oversegmentation and undersegmentation
Our probabilistic description allows us to add and merge nodes in the graph in a statistically rigorous manner to tackle the track fragmentation caused by oversegmentation and undersegmentation. The procedure relies on four key parameters: the false positive and false negative rates of the cell detection network and the predicted link and division probabilities of each cell. The false positive and negative rates follow automatically from the validation of the cell detection neural network that happens during the training phase, while the link and division probabilities are (automatic) predictions from the linking neural network and the division neural network. Hence, these parameters are in principle obtained through the network training procedure, without any further user intervention.
Oversegmentation occurs when a single cell generates two or more cell detections, potentially during multiple frames, causing tracks to split up erroneously. Such split tracks are identified as follows: these pairs of tracks should partially overlap in time (minimum of one frame and maximum of three frames) and nodes in the different tracks should be connected by relatively high-probability edges that are otherwise not part of the tracking solution. This reflects the fact that, if the tracks represent the same cell, edges between nodes in the two tracks should be likely. If the combined probability of an edge connecting the two tracks and the probability of a false positive cell detection, as given by the false positive error rate being higher than the probability of a track disappearing and another appearing (based on the false negative rates), we connect the tracks and prune the overlapping cell detections. We add a penalty, reflecting the false positive rate, to the energy of the link connecting the two tracks. This accounts for the fact that we have ignored a cell detection in creating the link (Extended Data Fig. 1a).
Undersegmentation occurs when a cell is not detected, leading to a single track becoming fragmented into two tracks. We identify fragmented tracks with a single frame gap between them and propose a new node that connects the tracks only if their start and end points are within a sufficiently short distance. Here, a cell detection is considered near to another one if it is one of the six closest neighbors. The added node receives a 3D position that is the average of the positions of the start and end points of the two tracks and is assigned a probability of being correct that is equal to the false negative error rate. In the graph containing all potential links, new edges are then made to all nearby nodes, with an energy penalty representing a uniform link probability (Extended Data Fig. 1b).
On a practical level, post-processing is implemented by first identifying all situations in which cell tracks appear or disappear. The algorithm then first addresses oversegmentations by attempting to connect appearing tracks with a nearby disappearing track that overlap in time for a maximum of three frames. After that, the algorithm addresses undersegmentation by attempting to connect appearing tracks with a nearby disappearing track that has disappeared just one time frame before.
Fundamentally, our post-processing solves a fundamental drawback of graph-based tracking frameworks that treat every cell detection as independent evidence for the existence of a cell. If, for instance, a cell is oversegmented in multiple subsequent time points, this is treated as very strong evidence that there are actually two cells present. It is obvious that this actually confers little more evidence than a single oversegmentation because these detections are and should be highly correlated between frames. Revisiting potential oversegmentations during post-processing allows us to treat multiple subsequent oversegmentations as a single false positive event.
Our undersegmentation correction method solves another problem with using flow solvers for tracking: they can ignore cell detections when making tracks but cannot add nodes for missed cell detections. A priori, it is difficult to determine where ‘helper’ nodes might need to be added, and allowing cell ‘merging’ to deal with undersegmentation18 makes the tracking problem much less constrained. We instead solve it with an easily understandable and straightforward post-processing step. Earlier cell-tracking solutions have employed conceptually similar methods but have to rely on manually picked parameters to regulate post-processing in the absence of a probabilistic description23. By contrast, we use our probabilistic graph description to rigorously identify the proper post-processing steps with minimal need for user-set parameters.
There is in principle no need for the user to adapt the post-processing procedure for different datasets, as long as the neural network-predicted probabilities are well calibrated and the user-set (dis)appearance probabilities are realistic (‘Graph description’). Similarly, retraining the neural networks for a new dataset automatically ensures proper post-processing on the new dataset as well. When reusing already trained neural networks in a new context, it can be beneficial to change the (dis)appearance probabilities to reflect the performance of the cell detection neural network in this new context.
Marginalization
Marginalization is performed on a subset of the graph to make it computationally tractable. We assume that the most informative edges (and their associated nodes) are between the same time points as the link of interest and are the ones closest to it in space. Distance is measured by how many steps on the graph have to be made to traverse edgewise from the target node of the link of interest (Extended Data Fig. 2b). Taking three steps as cutoff for inclusion in the subset yielded a computation time for marginalization similar to the time needed for neural network prediction of link and division probabilities, ~1 h for an imaging experiment of over 300 frames with over a hundred cell detections per frame.
The number of steps used to construct the subgraph can be changed by the user. We find that going beyond three steps, which already includes all the links of neighboring cells, does not meaningfully improve prediction quality (Extended Data Fig. 2c). This lack of improvement can partly be explained by marginalized link predictions for which the subgraph does not change when increasing from three to four steps (~30% of cases). For ~20%, the subgraph simply has no connections beyond three steps. For the remaining ~10%, the four-step subgraph has too many elements to be evaluated in a reasonable time and we are forced to use the three-step subgraph instead. Here, we use a cutoff so that no more than ~216 possible tracking solutions have to be checked. However, for links for which the subgraph used does grow, we still see little change in the prediction. This is mostly because many of the predictions with the three-step subgraph were already very high confidence (64% of cases are above 99.99% or below 0.001%), suggesting that most contextual information was already incorporated. Any further improvement thus made little difference for the prediction quality as measured by the cross-entropy loss.
For every node in the subset, all edges that point to nonmembers of the subset are combined in a single edge that accounts for the total probability to connect to a node outside of the set.
After subset selection, we construct a set of potential tracking solutions, test which solutions fit the graph constraints and calculate their associated energy. To avoid having to check the full set of binary combinations of events (~2N), we construct the set by varying, for every target node in the t + 1 time point, which node in the previous time point t it is connected to and combining all these variants. In this manner, the number of constructed potential solutions scales as \(\sim {({N}_{\rm{L}}/{N}_{\rm{T}}+1)}^{{N}_{\rm{T}}}+{2}^{{N}_{\rm{s}}}\), in which NT is the number of target nodes, Ns is the number of source nodes and NL is the number of edges. The target nodes can on average contact NL/NT possible source nodes and can appear without a source (first term), while the cells represented by the source nodes can either disappear or not (second term). We will refer to these possible variants as ‘microstates’ of which we will later combine the probabilities to compute the error rates.
Microstates can be encoded as a vector with its length as the number of events (1 if an event, such as a link or division, is part of it; 0 if not). To check if a microstate is possible, we can construct a matrix that encodes the flow constraints on the graph. This matrix gives the net flow into every node when multiplied with a microstate vector. An outgoing link or disappearance event represents a flow of −1, while an incoming link or appearance gives a flow of 1. Divisions are represented with a −1 flow, as they should allow an extra outgoing link. When for one or more of the nodes, the flow is unbalanced, the microstate is rejected and excluded from the partition function. Total energies are calculated by taking the inner product with a vector containing the energy penalty per event. These energies are then divided by the ‘temperature’ for proper calibration (‘Motivation for using ‘temperature scaling’’).
The probability of a link of interest (A) being true given all predictions made on the elements of the subgraph (G) is thus found by normalizing the probabilities associated with microstates containing that link to the sum of the probabilities of all possible microstates. The probability of a given microstate (WA) in turn is proportional to the exponent of the negative sum of the energy of all its elements (E(WA)):
To reduce the computational burden, links that are deemed almost certainly correct (>99.99%) or incorrect (<0.01%) are marginalized over a minimal subgraph containing only the other input edges of the target node of the link in question. When the estimated number of microstates that would need to be constructed exceeds 216, we shrink the subgraph by one ‘step’ to avoid long computation times.
Motivation for using ‘temperature scaling’
The marginalization procedure without temperature scaling assumes that the energy penalties are derived from information that is unique to the predictor, a neural network in our case. This is not a realistic assumption, as predictions might be made on the basis of overlapping crops and on shared baseline estimates. Not accounting for this overlapping information leads to overconfidence (Extended Data Fig. 2).
In our solution for this problem, we propose to split all predictions in a component that is based on inputs shared between neural networks and in one based on information unique to that prediction. The predictions (p) can then be seen as the product of the relative probabilities based on this shared and unique information:
This allows us in turn to split up the energy (the negative relative log likelihood) in a shared and unique component. We then assume that the energy related to the probability based on the shared inputs is proportional to the total energy (Ei). This assumption reflects our intuition that the confidence of neural network prediction should be reflected by both the unique and shared component. If, for instance, a link is highly likely, then this can probably be deduced both from the shared and the unique information available to the network and both energies should be highly negative. This gives:
where a is a constant between zero and one.
When calculating the energy of a microstate (\({E}_{W}^{* }\)), we can then sum the unique energies while assuming we can combine the shared information in a weighted manner. This weighing factor b (smaller then 1) should be low if all the shared information is shared between all events and higher if the overlap is less (for instance, when a prediction made about a link mostly shares information with adjacent links but not with all elements in the subset):
From this, we derive that we can account for shared information by using a single factor that functions as a temperature (T). This temperature is high if much of the information in any given prediction is not unique (high a) and if this shared information is shared with all other predictions (low b):
Marginalization as an opinion-pooling procedure
We can also motivate our marginalization procedure without relying on analogies with statistical physics. Instead, we can interpret our method as an extension of the ‘multiplicative opinion-pooling’ framework proposed by Dietrich24,25. The idea of combining predictions in a machine learning context has an older history56, but the specific framework of Dietrich and List enables us to neatly deal with prior probabilities and overlapping information. This will prove to be key in producing well-calibrated outputs.
Multiplicative opinion pooling suggests that opinions of different agents (different predictions by neural networks in our case) can be combined by multiplying them:
in which ω is a state in the set of possible state and Pi and denotes the probabilities predicted by individual predictors. Shared information between predictors can be incorporated in this framework by normalizing the predictions to the priors of the predictors based on the shared information. Conceptually, this means that predictors first arrive at a consensus P0 on the basis of their shared prior information, after which their unique information is pooled multiplicatively.
with c functioning as a normalization factor:
In this framework, each predictor must have an opinion on all possible states. In our case, predictors make only a single prediction on an event a (a link or division) that is part of a state. Therefore, we redefine multiplicative pooling as:
in which the microstate probability is now proportional to the product of the probabilities that its constitutive parts are true and the other events are false. The probability of a given event can then simply be calculated as:
By extending multiplicative pooling in this manner, we retain a major motivation behind multiplicative pooling, namely ‘individual-wise bayesianity’. This axiom states that it should not matter to the final prediction whether extra information is integrated before or after the pooling procedure, as the input information is the same. In our case, this holds on two levels (see Supplementary Discussion for the proof). First, it does not matter when we introduce information about a microstate when calculating its probability (P(ω)). It also does not matter when information about an individual event is introduced when we are calculating its probability (P(a)). This provides large flexibility in post hoc integration of new opinions, such as the judgment of a human reviewer.
The question remains of how to extend our concept of ‘temperature’ to this framework. For simplification, we can rewrite everything in terms of relative probabilities (Li = pi/(1 − pi)):
The question now remains of how to define the consensus prior L0 and determine the priors. Dietrich and List suggest using a geometric mean on the priors if the shared information is completely shared between all agents25. In our case, this is not necessarily true; therefore, we let the weight associated to a single prediction be free (b) instead of 1/n. For the priors, we again assume that the shared information is proportional (with a factor a) to the total information held by an agent.
which is equivalent to the description we arrived at using the statistical physics framework.
We finally wish to contrast this opinion-pooling procedure with updating a ‘Bayesian belief matrix’, a (cell-)tracking approach that uses link probability estimates to connect nondividing object detections32,57. This method cannot integrate division probabilities and can only take one type of constraint into account: the fact that cells cannot merge. In situations in which these are the only constraints present (for instance, when considering a subgraph in which only one cell is present at the later time point), we show that this approach is equivalent to our marginalization method (Supplementary Discussion).
Estimating the calibration temperature
We find the optimal temperature (as defined by the binary cross-entropy loss) by calibrating on the training data. To do this, we use neural networks to predict link and division probabilities for the cell detections in the training data. Next, we perform marginalization and compare marginalized link probabilities to the manual tracking. The task is now to find a ‘temperature’ (T), for which the predictions pi are closest to the ground truth (li denotes the truth value of a link). That is, the temperature for which the likelihood of the ground truth given the predictions is maximized, and the binary cross-entropy is thus minimized:
with pi given by:
In practice, most of the energy contribution in our marginalization comes from a handful of, often two, microstates. As an example, the dominant microstates of an uncertain link often take the form of an option in which all cells move half a cell to the left and another in which they move half a cell to the right. For a given link A, one of these states dominates the microstates that contain the link (WA) and the other state dominates the ones that do not contain it (W\A). This allows us to approximate the marginalized probability pi,T as:
This clearly and conveniently maps on a linear regression problem, for which we have to learn parameter 1/T given the original marginalized relative likelihoods (Li,T = 1) as an input.
The temperature obtained in this manner works well on data that are part of (Extended Data Fig. 2) and outside (‘Evaluation of the marginalization procedure’) the training dataset in producing well-calibrated error rates. This proves that the simplifications made to arrive at a single correction factor amenable to linear regression are allowable.
The obtained temperature (similar to the Platt scaling parameters previously) is used in our algorithm as a point estimate without considering the uncertainty associated with the calibration. This can be justified by the tight confidence intervals we obtain (Extended Data Fig. 2d) and the robust calibration we see across datasets (Fig. 3 and Extended Data Fig. 3). For a fully Bayesian description of our framework, which includes calibration uncertainty, see the Supplementary Discussion (Full Bayesian description of link error prediction framework).
Evaluation of the marginalization procedure
To evaluate the correctness of the marginalized error rates, we again compare our predictions against the five fully manually annotated organoids used for the other evaluations. We use the manually annotated cell centers as the input for our division and link detection and perform marginalization afterward. This allows us to compare all error rate estimates to a fully human-derived ground truth, without the need to map machine-predicted cell centers on the cell centers annotated by humans. These mappings are not trivial, and errors in these mappings can strongly skew the results. Furthermore, because the human-assigned links are completely independent from the algorithm output, we deem this the strongest test for our marginalization procedure.
We bin the marginalized link predictions in groups based on their relative log likelihood (15 bins). For every bin, we then compute the average probability of the link being correct and compare this to the actual amount of the correct link in this bin as determined by examining the ground truth.
We also perform this evaluation on the manually reviewed tracking data (‘Manual review’). Here we started out with cell centers predicted by a neural network. Verifying that the error rates are well calibrated in this case shows that the marginalization procedure is not dependent on human-annotated cell centers. We compare the error rates against tracking data when all links are corrected but no undersegmentations or oversegmentations are fixed (Extended Data Fig. 3). Fixing segmentation errors involves changing the graph representation and thus introduces links without an associated error rate prediction, making evaluation impossible.
Manual review
To evaluate manual annotation, we reviewed the possible errors for three complete organoids tracked for around 100 to 300 frames. Potential errors were flagged at all links that had a marginalized probability below 99% and the start and end points of appearing and disappearing tracks, respectively. We first corrected all potential link errors and used the corrected data to check the calibration of the marginalized predictions as described above. We then checked all other errors and identified their cause for the largest (>300-frame-long) dataset.
Error correction was carried out in our GUI4, which zooms in on errors and informs user about the kind of error they encountered: possible link mistake, track appearing or track disappearing. The GUI also allows backtracking, that is, the selection of cells of interest, based for instance on their cell type or final position, to focus curation and analysis only on these cells.
Palbociclib intestinal organoid tracking
Palbociclib (at a final concentration of 10 μM) was added 2 d after seeding, and organoids were then imaged for 2 d. Three crypts that stayed in the field of view for the full image duration were chosen for analysis.
To create the ground truth dataset, 50 frames each in two organoids were manually corrected around 20 h after palbociclib treatment.
Out-of-sample use: image preprocessing
For out-of-sample usage of our intestinal organoid trained neural network, we have identified two key preprocessing steps to improve tracking results: scaling and background subtraction. First, regarding scaling, CNNs (and UNETs by extension) are generally not scale free. Therefore, to avoid oversegmentation or undersegmentation, the nuclear size should match the nuclear sizes in the training data. For same-sized nuclei imaged on different microscopes, this typically corresponds to matching the pixel resolution. Second, during the acquisition of the intestinal organoid training data, the detector gain and offset was set such that the background (meaning the region outside the organoid) largely had fluorescence values of zero. Subtracting the background so that this holds for the out-of-sample dataset as well helps to restrict the cell detections to the region containing the tissue, reducing false positive cell detections. Lastly, we have seen that, for data in which cells have large differences in nuclear fluorescence (unpublished), it helps to reduce the contrast using a gamma correction.
Out-of-sample use: recalibration
Recalibration of the error rates for out-of-sample data follows the same process as the initial estimation of the scaling temperature (‘Estimating the calibration temperature’): we compare the marginalized predictions against a ground truth dataset to find the optimal temperature that minimizes the cross-entropy loss between predictions and truth values.
To create the ground truth, the user has to correct potential mistakes in a number of representative frames. We find that correcting around 200 potential linking mistakes is generally enough to obtain tight estimates of the new scaling temperature (Extended Data Fig. 7e,f). The procedure requires users to review all potential errors (<99% probability) in a given frame to avoid bias in which mistakes are corrected.
The recalibration procedure thus functions as follows:
-
1.
Predict tracks and compute error rates.
-
2.
In the manual curation GUI, select frames in which to correct potential mistakes (aim for more than 200 potential mistakes).
-
3.
Correct mistakes in the GUI.
-
4.
Recalibrate the error rates using the temperature-scaling functionality.
-
5.
Recompute the error rates with the new scaling temperature.
A graphic description of the pipeline users have to follow when using neural networks on out-of-sample data including both the image-preprocessing and recalibration steps can be found in Extended Data Fig. 10.
Out-of-sample use: low-SNR intestinal organoid tracking
The low-SNR intestinal data were taken (but not yet analyzed) in the context of Zhang et al., and imaging was carried out as described in their paper6. Preprocessing consisted of downscaling in xy by a factor of 1.33 to correct for different pixel resolution and background subtraction using a tophat filter. Post-processing was changed to retain deep tracks, up to 60 µm deep in the tissue.
Out-of-sample use: blastocyst tracking
The two longest time series in the BlastoSPIM dataset29 were chosen for analysis (the series starting with F30 and F41, respectively). Preprocessing consisted of downscaling the image in xy by a factor of 2.5 and background subtraction by subtracting a constant value. This downscaling was not meant to match the pixel resolution of the blastocyst data to that of the training data (different by a factor of 1.25 in xy) but rather to match the nuclear volumes, which are considerably larger in the blastocyst (nuclear radius29 of ~6.5 µm versus ~3.5 µm in intestinal organoids). When a blastocyst underwent a major rotation, we ignored that time point in our analysis of the error rates. These major rotations occurred only during two frames in only one of the blastocysts. To correctly track cells through these major rotations, our tracking algorithm would simply have to be combined with an image registration step, as in the original paper describing the dataset29.
Out-of-sample use: C. elegans cell tracking
We obtained the C. elegans embryo datasets from the Cell Tracking Challenge website17. The available training data consisted of two fully annotated movies (~150 frames long) following cells from two-cell to ~128-cell stages.
We trained new cell detection and link and division prediction neural networks on the two provided annotated training datasets. As all cells in the imaging volume were annotated, there was no need to crop the image during cell detection training, and we could increase the background weighting to 0.95 without risking training on unannotated cell centers. Due to the difference in nucleus sizes compared to those of the intestinal organoid data, we also increased the radius parameters in the distance mapping for cell detection. All other networks were trained as for the intestinal organoids. We estimated the proper scaling temperature for the marginalization by calibrating on the training data as described for the intestinal organoid data.
We used one of the unannotated ‘challenge’ datasets to evaluate tracking quality and validate that the marginalized probabilities were well calibrated. We did this by manually checking all potential errors and using the corrected dataset as our reference.
Automated lineage dynamics analysis
For the automated analysis, we first filtered out all links that had a marginalized probability below 99%. All tracks that do not end in a division are considered censured.
A key assumption underlying survival analysis is that the probability of an event happening is independent of the chance of being lost to follow-up. In our case, this assumption is broken, as cells are relatively often lost when they are close to dividing (due to rapid nucleus movement) and cell division is the key event when studying lineage dynamics. This means that we would underestimate the number of dividing cells, because we tend to lose track of them just before they divide. We break this dependency by using a division detection neural network to check for every track that is lost to follow-up if it is lost during the division process. We then reassign tracks that end in a predicted division (>50% predicted probability) from the censured category to the divided class. Now observing the division events is no longer affected by uncertainties in tracking during the division process.
The neural network trained for this task was trained in the exact same manner as described before except that we are not interested in pinpointing the exact moment of chromosome separation. We also wish to classify tracks as dividing if they are lost during any other moment of the division process. We therefore classify all cells within two frames around division as dividing during training. Because of the varying length of the division process, we exclude time points directly around this window to avoid including cells in the training data that look clearly mitotic but are just outside the window.
We can also use this neural network to split tracks that contain a division but were not assigned as dividing in initial tracking due to lack of a plausible daughter cell, for instance, because one of the daughters moved out of view. Therefore, we break up tracks when the chance of division is on average higher than 99% for three consecutive frames.
Our method detects some cells with very short cell cycles, in which cell division generally leads to cell death and not in two daughter pairs, potentially reflecting polyploid cells. These are not classified as dividing in the manually annotated data; therefore, we remove these very short cell cycles (less than 6 h). This has the added benefit that it also removes some cases in which the division neural network wrongly assigns a division to a track end. Although the chance of this happening is low, it happens generally in less than 2.5% of tracks.
For survival analysis, we use the ‘surv’ package in R and for the fitting ‘survflexcure’. During analysis, we only use tracks that start in a division and use the next division as the event under study. Cell cycle times are analyzed by fitting a Guassian hazard to the data, allowing for a ‘cured’ fraction that will not divide again. The mean of the Gaussian represents the average cell cycle, and its standard deviation represents the spread around this mean. The ‘cured’ fraction is used as an estimate of the fraction of differentiating cells. Before fitting, we remove outliers that are more than ~7 h from the mean (more than three times the standard deviation). To avoid dealing with negative times, we fitted a log normal distribution to the exponents of the survival times instead of using a normal distribution directly.
Manual data for comparison are analyzed in the same manner, but the only censoring events derive from cell death, the end of the experiment or cells leaving the imaging volume. No neural network thus has to be used to check whether censured tracks end in a division.
Video visualization
Three-dimensional rendering of the microscopy with overlaid tracks was carried out using Napari. We have written a plugin (available on GitHub) that allows importing of tracking results into Napari. The 3D reconstruction of lineages using manually curated data (Supplementary Video 4) was carried out using ParaView. We provide extensive documentation that can be used to reproduce this visualization.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Due to storage considerations, all imaging data and accompanying tracking data are available upon request. Sample imaging and tracking data are available on Zenodo (https://doi.org/10.5281/zenodo.13982844)58.
Code availability
OrganoidTracker software is freely downloadable from GitHub (https://github.com/jvzonlab/OrganoidTracker). The models used are available on Zenodo (https://doi.org/10.5281/zenodo.13912686, https://doi.org/10.5281/zenodo.13946119)59,60.
References
Betjes, M. A., Zheng, X., Kok, R. N. U., van Zon, J. S. & Tans, S. J. Cell tracking for organoids: lessons from developmental biology. Front. Cell Dev. Biol. 9, 675013 (2021).
Beumer, J. & Clevers, H. Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol. 22, 39–53 (2021).
de Medeiros, G. et al. Multiscale light-sheet organoid imaging framework. Nat. Commun. 13, 4864 (2022).
Kok, R. N. U. et al. OrganoidTracker: efficient cell tracking using machine learning and manual error correction. PLoS ONE 15, e0240802 (2020).
Huelsz-Prince, G. et al. Mother cells control daughter cell proliferation in intestinal organoids to minimize proliferation fluctuations. eLife 11, e80682 (2022).
Zheng, X. et al. Organoid cell fate dynamics in space and time. Sci. Adv. 9, eadd6480 (2023).
He, Z. et al. Lineage recording in human cerebral organoids. Nat. Methods 19, 90–99 (2022).
Ender, P. et al. Spatiotemporal control of ERK pulse frequency coordinates fate decisions during mammary acinar morphogenesis. Dev. Cell 57, 2153–2167 (2022).
Sugawara, K., Çevrim, Ç. & Averof, M. Tracking cell lineages in 3D by incremental deep learning. eLife 11, e69380 (2022).
Malin-Mayor, C. et al. Automated reconstruction of whole-embryo cell lineages by learning from sparse annotations. Nat. Biotechnol. 41, 44–49 (2023).
Wait, E. et al. Visualization and correction of automated segmentation, tracking and lineaging from 5-D stem cell image sequences. BMC Bioinformatics 15, 328 (2014).
Bragantini, J., Lange, M. & Royer, L. A. Large-scale multi-hypotheses cell tracking using ultrametric contours maps. Preprint at https://arxiv.org/abs/2308.04526 (2023).
Mitrophanov, A. Y. & Borodovsky, M. Statistical significance in biological sequence analysis. Brief. Bioinform. 7, 2–24 (2006).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Soneson, C. & Delorenzi, M. A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics 14, 91 (2013).
McKinley, K. L. et al. Cellular aspect ratio and cell division mechanics underlie the patterning of cell progeny in diverse mammalian epithelia. eLife 7, e36739 (2018).
Maška, M. et al. The Cell Tracking Challenge: 10 years of objective benchmarking. Nat. Methods 20, 1010–1020 (2023).
Haubold, C., Aleš, J., Wolf, S. & Hamprecht, F. A. A generalized successive shortest paths solver for tracking dividing targets. in Computer Vision — ECCV 2016 (eds Leibe, B. et al.) 566–582 (Springer International, 2016).
Platt, J. C. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. in Advances in Large-Margin Classifiers (eds Smola, A. J. et al.) (MIT, 2000).
Guo, C., Pleiss, G., Sun, Y. & Weinberger, K. Q. On calibration of modern neural networks. Proc. Mach. Learn. Res. 70, 1321–1330 (2017).
Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T. & Ronneberger, O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 424–432 (Springer, 2016).
Mazzaferri, J., Roy, J., Lefrancois, S. & Costantino, S. Adaptive settings for the nearest-neighbor particle tracking algorithm. Bioinformatics 31, 1279–1285 (2014).
Löffler, K., Scherr, T. & Mikut, R. A graph-based cell tracking algorithm with few manually tunable parameters and automated segmentation error correction. PLoS ONE 16, e0249257 (2021).
Dietrich, F. Bayesian group belief. Soc. Choice Welf. 35, 595–626 (2010).
Dietrich, F. & List, C. in the Oxford Handbook of Probability and Philosophy (eds Hájek, A. & Hitchcock, C.) (Oxford University Press, 2016).
Dey, T. et al. Survival analysis—time-to-event data and censoring. Nat. Methods 19, 906–908 (2022).
Aspert, T., Hentsch, D. & Charvin, G. DetecDiv, a generalist deep-learning platform for automated cell division tracking and survival analysis. eLife 11, e79519 (2022).
Mohammadi, F. et al. A lineage tree-based hidden Markov model quantifies cellular heterogeneity and plasticity. Commun. Biol. 5, 1258 (2022).
Nunley, H. et al. Nuclear instance segmentation and tracking for preimplantation mouse embryos. Development 151, dev202817 (2024).
Hirsch, P. et al. Tracking by weakly-supervised learning and graph optimization for whole-embryo C. elegans lineages. In International Conference on Medical Image Computing and Computer-Assisted Intervention 25–35 (Springer, 2022).
Murray, J. I. et al. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat. Methods 5, 703–709 (2008).
Ulicna, K., Vallardi, G., Charras, G. & Lowe, A. R. Automated deep lineage tree analysis using a Bayesian single cell tracking approach. Front. Comput. Sci. 3, https://doi.org/10.3389/fcomp.2021.734559 (2021).
Turetken, E., Wang, X., Becker, C., Haubold, C. & Fua, P. Network flow integer programming to track elliptical cells in time-lapse sequences. IEEE Trans. Med. Imaging 36, 942–951 (2016).
Moen, E. et al. Accurate cell tracking and lineage construction in live-cell imaging experiments with deep learning. Preprint at bioRxiv https://doi.org/10.1101/803205 (2019).
Wen, C. et al. 3DeeCellTracker, a deep learning-based pipeline for segmenting and tracking cells in 3D time lapse images. eLife 10, e59187 (2021).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Villars, A., Letort, G., Valon, L. & Levayer, R. DeXtrusion: automatic recognition of epithelial cell extrusion through machine learning in vivo. Development 150, dev201747 (2023).
Roudot, P. et al. u-track3D: measuring, navigating, and validating dense particle trajectories in three dimensions. Cell Rep. Methods 3, 100655 (2023).
Arbelle, A., Reyes, J., Chen, J.-Y., Lahav, G. & Riklin Raviv, T. A probabilistic approach to joint cell tracking and segmentation in high-throughput microscopy videos. Med. Image Anal. 47, 140–152 (2018).
Tian, C., Yang, C. & Spencer, S. EllipTrack: a global–local cell-tracking pipeline for 2D fluorescence time-lapse microscopy. Cell Rep. 32, 107984 (2020).
Magnusson, K. E., Jalden, J., Gilbert, P. M. & Blau, H. M. Global linking of cell tracks using the Viterbi algorithm. IEEE Trans. Med. Imaging 34, 911–929 (2015).
Krotenberg Garcia, A. et al. Active elimination of intestinal cells drives oncogenic growth in organoids. Cell Rep. 36, 109307 (2021).
Basak, O. et al. Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell 20, 177–190 (2017).
Verissimo, C. S. et al. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. eLife 5, e18489 (2016).
Barbáchano, A. et al. Organoids and colorectal cancer. Cancers 13, 2657 (2021).
Lukonin, I., Zinner, M. & Liberali, P. Organoids in image-based phenotypic chemical screens. Exp. Mol. Med. 53, 1495–1502 (2021).
Stringer, C. & Pachitariu, M. Cellpose3: one-click image restoration for improved cellular segmentation. Nat. Methods 22, 592–599 (2025).
Kok, R. N. U., Spoelstra, W. K., Betjes, M. A., van Zon, J. S. & Tans, S. J. Label-free cell imaging and tracking in 3D organoids. Cell Rep. Phys. Sci. 6, 102522 (2025).
O’Connor, O. M. & Dunlop, M. J. Cell-TRACTR: a transformer-based model for end-to-end segmentation and tracking of cells. PLoS Comput. Biol. 21, e1013071 (2025).
Gallusser, B. & Weigert, M. TRACKASTRA: transformer-based cell tracking for live-cell microscopy. in Computer Vision — ECCV 2024 (eds Leonardis, A. et al.) 467–484 (Springer Nature, 2024).
Hastings, W. K. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57, 97–109 (1970).
Metropolis, N., Rosenbluth, A. W., Rosenbluth, M. N., Teller, A. H. & Teller, E. Equation of state calculations by fast computing machines. J. Chem. Phys. 21, 1087–1092 (1953).
McDole, K. et al. In toto imaging and reconstruction of post-implantation mouse development at the single-cell level. Cell 175, 859–876 (2018).
Höfener, H. et al. Deep learning nuclei detection: a simple approach can deliver state-of-the-art results. Comput. Med. Imaging Graph. 70, 43–52 (2018).
Liu, R. et al. An intriguing failing of convolutional neural networks and the coordconv solution. In Proc. of the 32nd International Conference on Neural Information Processing Systems (eds Bengio, S. et al.) 9628–9639 (Curran Assoc., 2018).
Hinton, G. E. Product of experts. In 1999 Ninth International Conference on Artificial Neural Networks ICANN 99. (Conf. Publ. No. 470) 1–6 (IET, 1999).
Narayana, M. & Haverkamp, D. In 2007 IEEE Conference on Computer Vision and Pattern Recognition 1–8 (IEEE, 2006).
Betjes, M. Example data OrganoidTracker 2.0. Zenodo https://doi.org/10.5281/zenodo.13982844 (2024).
Betjes, M. Example data OrganoidTracker 2.0. Zenodo https://doi.org/10.5281/zenodo.13912686 (2024).
Betjes, M. Models trained on intestinal organoid data. Zenodo https://doi.org/10.5281/zenodo.13946119 (2024).
Acknowledgements
We thank G. Huelsz-Prince and X. Zheng for their work in the creation of annotated intestinal organoid datasets. This work is funded by the ‘Organoids in time’ grant (OCENW.GROOT.2019.085, M.A.B.) and the Building Blocks of Life grant (737.016.009, R.N.U.K.) from the Netherlands Organization for Scientific Research (NWO).
Author information
Authors and Affiliations
Contributions
Conceptualization: M.A.B., S.J.T. and J.S.v.Z.; methodology: M.A.B.; data curation and annotation: M.A.B.; coding: M.A.B. and R.N.U.K.; GUI: M.A.B. and R.N.U.K.; visualization: M.A.B.; writing: M.A.B., S.J.T. and J.S.v.Z.; supervision: S.J.T. and J.S.v.Z.; funding acquisition: S.J.T. and J.S.v.Z.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Sundar Naganathan, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Rita Strack, in collaboration with the Nature Methods team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Post-processing of the tracking solution.
a) Illustration of how post-processing deals with over-segmentations. The blue links represent the maximum-likelihood solution, while the grey links are excluded from the solution. Over-segmentations are not independent events, but are treated as such by the min-cost flow solver, which can lead to fragmentation of tracks (left panel). During post-processing we check if track fragments can be joined by adding a single link and removing the cell detections associated with the over-segmentation. Removing multiple of these nodes is associated with a single penalty (Eignore, the relative log-likelihood of over-segmentation, based on the false positive cell detection rate) which is added to the new link so that our probabilistic description remains correct. b) Illustration of how post-processing deals with under-segmentations. The min-cost flow solver cannot add nodes to the graph, which can lead to fragmentation of tracks (left panel). During post-processing we check if track fragments can be joined by adding a node. Adding a node is associated with a penalty (Eaddition, the relative log-likelihood of missing a cell detection, based on the false negative detection rate) and the node is connected to nearby nodes in the graph with uniform probability. c) Error-free track length distributions in a representative tracked organoid. The ground truth (green) represents fully manually corrected data, where tracks are only cut short by cell death, the end of the experiment, or leaving the field-of-view. The initial maximum-likelihood solution (gray) has many more short tracks, which is partly solved by post-processing (blue). d) The fraction of cells present at a certain time that can be tracked without error from the start of the experiment. Only cells that are trackable for the full experiment in the ground truth are considered. By post-processing the data we can increase the fraction of cells that are trackable for the complete experiment (>60 h) from around 50% to around 75%. See the section ‘Solving over- and undersegmentation’ in the Methods for more details.
Extended Data Fig. 2 Temperature scaling during the marginalization procedure.
a) Visual illustration of the temperature scaling procedure. The prediction on a single link (encoded in its weight) is not independent from predictions on nearby links. To account for this shared information we divide the link energy by a ‘calibration temperature’ (Eq. 1). The marginalized link probability, P(A|B), is then based on scaled energies (Eq. 2). The calibration temperature is found by minimizing the cross-entropy loss (CE) between the marginalized predictions and the ground truth (y), with respect to this temperature (Eq. 3). This scaling temperature has to be calibrated only once after training a set of division and link neural networks. The calibration can be done on the same data that was used for neural network training. For data far outside the training distribution new calibration can be performed on a small manually corrected set of links. b) Illustration of the difference subsets used in the subsequent plots. Red symbolizes the naïve approach, where the subset simply is the link of interest. In this case no marginalization is done. The green subset only considers link at a distance of one step from the target node of the link of interest. The blue subset goes up to a distance of three steps. This is the largest set that is computationally feasible (~1 h of computation time for 300 frames). c) Prediction performance as measured by the binary cross entropy loss versus the subgraph size, all dots are individual organoids. The loss at the optimal temperature (lowest point in panel d) was taken. Increasing subgraph size improves prediction, but there is no improvement when going beyond three steps, see method section ‘Marginalization’ for further discussion. d) Cross entropy loss between predictions and ground truth (based on all 9 organoids in the training dataset) for different neighborhoods and temperatures. Minimum loss (dotted line) is achieved at higher temperatures when the subset gets larger. All lines represent individual organoids. e) Optimal calibration temperature for every neighborhood. The error bars represent the 95% confidence interval (this is a lower bound as not all observed links are truly independent). Limiting ourselves to calibration on the validation dataset that was left during training gives the same results. f) Optimal temperatures and confidence intervals (again lower bounds) per organoid. The estimates show great overlap between organoids, validating that we can use a single calibration temperature for all. g) Predicted versus actual log-likelihoods after marginalization on different subsets. When marginalizing on larger subsets predictions become overconfident. If predictions suggest that links are not correct, more than expected fraction is actually correct and the other way around. h) After scaling the energies with the proper temperature per subset, we get well-calibrated link predictions.
Extended Data Fig. 3 Marginalization during full procedure.
a) The predicted relative log10-likehoods from the link neural network (black line) is well-calibrated when compared to the manually corrected ground truth (dotted line denotes perfect calibration). The overestimation at low likelihoods is due to the filtering of unlikely links before tracking and marginalization (see Methods). This filtering preferentially removes low-probability incorrect links, making the remaining ones more likely to be true. The fact that links in the global solution (blue line) are much more likely than expected and links excluded from the tracking solution (gray line) are less likely than expected, suggests that contextual information could improve the error rate prediction. b) Many links in the tracking solution are less than 99% (black line) certain based on the naïve predictions. c) Marginalization integrates context and largely removes the discrepancy between links in and out of the tracking solution. d) Only few links in the tracking solution are less than 99% certain after marginalization. e) The fraction of uncertain links (<99% certainty) as fraction of total. Marginalization decreases the amount of uncertain links around four-fold. The longest experiments, which have poorer signal to noise, benefitted most from the marginalization (the black lines denote individual experiments). The red fraction indicates actual errors in the low-confident fraction. The dotted line the fraction of errors across all links including ones that have a high probability of being true, showing that no significant amount of errors is missed. f) Five randomly selected lineage trees colored by their naïve predicted error rate (yellow to red links are uncertain. g) The same trees after marginalization.
Extended Data Fig. 4 Track error rates.
a) The predicted likelihood (after marginalization) of links in the tracking solution against the measured likelihood of being correct based on the manual data (top panel). The line does not overlap the dotted line, because not all graph information available to the flow-solver can be integrated in the error prediction. Key is that they remain above the dotted line so that the error predictions are conservative. The histogram (bottom panel) shows the distribution of likelihoods of correct (green) and incorrect links (red). Because the linking error rate is so low, incorrect instances cannot be seen in the histograms a) and b). The black vertical line indicates the threshold of 99% chance of being correct. b) The likelihood of a pair of links both being correct can be calculated by combining their constituent probabilities by simple multiplication. It does not matter if the links are subsequent (blue line) or unconnected (gray line). It is thus not so that a link being true is informative of the truth of the subsequent link, beyond its predicted error probability. c) The predicted error rates for tracks of arbitrary length. The probability that a cell can be correctly traced back to its last division (red dotted lines) is predicted for every cell at every timepoint. The probability of the track being correct is calculated by multiplying all the constituent probabilities. The tracks are compared to the ground truth and deemed correct if they recapitulate it exactly, yielding a similar calibration curve to a). d) The same as in c) but now only with tracks that span the full cell cycle, again producing a similar calibration curve as in a). Error detection works efficiently for full tracks spanning the complete cell cycle, only one incorrect track is above the 99% certainty cut-off.
Extended Data Fig. 5 Survival curves for different organoids.
a) Kaplan-Meier curves describing the fraction of non-divided cells as a function of time since division, for 5 organoids where manual reference data was available for the complete crypt (grey curve). The shaded region denotes the 95% confidence interval. b) Kaplan-Meier curves describing the fraction of non-divided cells as a function of the time since the division of the sister cell. c) Comparison of manually annotated data to automated analysis for the estimation of three key parameters of lineage dynamics. Color indicates whether the variation in cell cycle duration or the probability that the cell will divide again was calculated for all cell cycles (red) or for cell cycles relative to the moment of division of the sister cell (blue). Cell cycle times show almost perfect correlation between manual and automated data. The deviation in the cell cycle time is more sensitive to outliers and consequently shows poorer correlation. The non-dividing fraction again shows strong correlation. d) Overlay of all Kaplan-Meier curves of automatically tracked organoids, n = 20, which describes the fraction of non-divided cells as a function of time since division. Black line represents the combined data. e) Same but as a function of time since the sister division. f) Statistical analysis of organoid lineage parameters. Error bars indicate standard deviations around the mean. Cell cycle times only show limited variation across organoids and experiments, with standard deviation <10% of the mean cell cycle time (left panel). The variation around the cell cycle mean for cell cycle times is significantly larger than the variation between the cell cycle times of sisters (middle panel), suggesting strong correlation between sister pairs. This is further supported by the results that given that the sister divides, the non-dividing fraction becomes close to zero instead of around 30% (right panel).
Extended Data Fig. 6 Tracking performance on organoids with perturbed dynamics.
(a) The number of detected cells in the imaging volume for two organoids treated with palbociclib, which blocks cell division. After ~10 h of Palbociclib exposure, cell death dominates over cell division, leading to decreased cell numbers. (b) Distribution of cell cycle lengths (0 h is cell cycle start) plotted for the same two organoids. Cells divide either slowly (>25 h) or not at all, consistent with Palbociclib action. (c) Thirty randomly selected lineages show tracking with very low potential error rates. Cells generally divide only once, or not at all. (d) Top: measured link likelihood versus likelihoods predicted by the neural network. Data is shown for all possible links (black) or links that are either in the global solution (blue). Dotted line is perfect calibration. Data is for the 2 different organoids in (a) and (b). Shaded region is S.D.M. Predictions for Palbociclib-treated organoids remain well-calibrated, indicating that perturbation of cell dynamics does not affect the quality of error predictions. Bottom: histogram of the predictions. Almost all links in the tracking solution (blue) are above the 99% probability threshold (vertical line) indicating highly confident tracking.
Extended Data Fig. 7 Performance for different image acquisition parameters.
(a) Tracking performance for intestinal organoids imaged on a different scanning confocal microscope (Leica TCS SP8) than used for collecting the training data (Nikon A1R MP). Consequently, imaging data had lower planar resolution (0.4 μm/px rather than 0.32 μm/px), cells could be imaged deeper into the organoid, but at low signal-to-noise. Green squares are tracked cells at 60 μm depth, while red and blue squares are their previous and next locations, respectively. The cells marked with an asterisk (*) correspond to cells in the lineage trees (right). Open circles (o) denote missed cell detections. Even at this comparatively low signal-to-noise ratio most (>80%) cells are detected. The YZ-cross-section show the depth of these cells in terms of cell number. Lineage trees are color-coded by predicted error rate. Most cells can be tracked for multiple hours without potential mistakes. b) Same as in a, but for cells at 50 μm. Signal-to-noise ratios are higher here and all cells are detected, while lineages show cell tracking for long (>10 h) periods without potential mistakes. c, d) Top: measured link likelihood versus likelihoods predicted by the marginalization procedure either without (c) or with (d) recalibration on newly corrected data. Data is shown for all possible links (black) or links that are either in the global solution (blue). Dotted line is perfect calibration. Data is for the 4 different data sets shown in (e) and (f). Shaded region is S.D.M. Without recalibration (c), predicted likelihoods are overconfident, for example the 99% probability threshold actually corresponds to a lower level of certainty. Using the actual 99% probability threshold would increase the number of links needed to be checked (c, bottom histogram). However, the number of flagged potential mistakes omitted when using the uncalibrated threshold remains low (0.06% of links). e) Visualization of the recalibration process. The recalibration constant, defined as the ratio of the old and new scaling temperature, is estimated by manual error correction of predicted tracks. Graphs show the estimated recalibration constant versus the number of frames corrected for four different data sets, as well as all data pooled. The estimates converge on the consensus estimate (dotted line) for >5 corrected frames. Shaded area denotes 95% confidence interval. f) The number of reviewed potential errors (<99% certainty links) as a function of frames corrected. Tight estimates of the recalibration constant are reached after ~200 reviewed potential errors. g) Survival curve indicating the probability that a cell has not divided at time t after its birth. Shown are the original (blue, green) and recalibrated (orange, red) data for the same organoid. h) Same as g) but for the timing of division relative to the sister division. The strong overlap between original and recalibrated data indicates that for downstream applications perfect calibration is often not essential.
Extended Data Fig. 8 Performance on different system: blastocyst.
(a) Lineages color-coded by error-rates for a blastocyst developing from the 16 to 64-cell stage (BlastoSPIM 1.0 dataset), as obtained by tracking without manual curation. Many cells can be tracked from the beginning to the end without any potential mistakes. b) Blastocyst cells undergoing large movements, with subsequent timepoints shown in green and magenta. Green square and red circles indicate the current and subsequent detected centroid position, respectively. The red line connects these positions, while the blue line connects to the centroid position in the previous time point. When either marker is not present, then the corresponding positions are >4 μm away from the z-slice shown. Our method tracks cells even for displacements larger than the typical nucleus size. c) Histogram of potential cell displacements in a single frame, for all potential links, with the correctness of each link established by manual curation. Many true links represent fast-moving cells, corresponding to displacements between timesteps of >1 cell radius, or ~6 μm, large compared to cell movements in intestinal organoids both in absolute and relative terms. d) Precision, recall and accuracy versus displacement between timesteps, for the optimal tracking solution before manual curation. Even fast-moving cells are tracked with high accuracy. e, f) Top: measured link likelihood versus likelihoods predicted by marginalization, either without (e) or with (f) recalibration on newly corrected data. Data is shown for all possible links (black) or links that are either in the global solution (blue). Dotted line is perfect calibration. Data is for 2 different blastocysts, corresponding to the data in (c) and (d). Shaded region is S.D.M. Without recalibration (c) predicted likelihoods are overconfident. Using a recalibrated threshold does not majorly increase the number of links to be checked (d, bottom histogram), as the bulk of links in the tracking solution remains above the 99% probability threshold (black vertical line).
Extended Data Fig. 9 C. elegans tracking.
a) Z-slices of 3D confocal data for C. elegans embryogenesis (training data). b) Using retrained neural networks, we obtain well-calibrated link-likelihood predictions (top panel). Almost all links in the tracking solution that minimize the global energy (blue) have very high marginalized likelihoods (bottom panel). In this data set, only few links in the graph are not in the tracking solution (grey) as the nuclei are less closely packed then in intestinal organoid data. c) Lineage trees with associated error rates show that only few potential errors (41 links with an error probability above 1%) are present after automated tracking. d) Lineage trees after manual correction of potential errors. The lineages map exactly on the known C. elegans AB, e, MS, c, d and P4 sub-lineages, strongly suggesting that no errors remain. e) Tracking statistics show a very low error rate and that almost no error in linking. Most errors arise from undetected nuclei (under-segmentation), deep in the imaging volume.
Extended Data Fig. 10 OrganoidTracker 2.0 workflow.
Pipeline for using already trained neural networks on new (out-of-sample) data.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10 and Discussion.
Supplementary Video 1
Tracking of intestinal organoids, cross-section. A single confocal image plane is shown over 60 h. All tracks up to 5 μm above or below this plane are plotted.
Supplementary Video 2
Tracking of intestinal organoids, 3D reconstruction. A 60-h-long 3D reconstruction of the intestinal organoid seen from the side closest to the objective, with all tracks overlaid.
Supplementary Video 3
Tracking of intestinal organoids, 3D reconstruction. A 60-h-long 3D reconstruction of the intestinal organoid seen from the side, with all tracks overlaid.
Supplementary Video 4
Tracking of intestinal organoids, corrected data. Left, depth-colored representation of the intestinal organoid data (60 h long). Right, 3D reconstruction of the intestinal organoid where every cell is represented by a sphere on the right, and five randomly chosen lineages are colored.
Supplementary Video 5
Organoid with low SNR, 50-μm depth. A single confocal image plane is shown over 40 h. All tracks up to 5 μm above or below this plane are plotted.
Supplementary Video 6
Organoid with low SNR, 60-μm depth. A single confocal image plane is shown over 40 h. All tracks up to 5 μm above or below this plane are plotted.
Supplementary Video 7
Blastocyst tracking, blastocyst 1. A 3D reconstruction of 25 h of mouse blastocyst light-sheet imaging data (BlastoSPIM dataset) with all the tracks overlaid.
Supplementary Video 8
Blastocyst tracking, blastocyst 2. A 3D reconstruction of 25 h of mouse blastocyst growth (BlastoSPIM dataset) with all the tracks overlaid. At two time points, the entire blastocyst undergoes a major rotation during which cells are not tracked.
Supplementary Video 9
C. elegans tracking. A 3D reconstruction of 5 h of confocal imaging data of C. elegans embryogenesis from the Cell Tracing Challenge with all the tracks overlaid.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Betjes, M.A., Kok, R.N.U., Tans, S.J. et al. Cell tracking with accurate error prediction. Nat Methods 22, 2400–2410 (2025). https://doi.org/10.1038/s41592-025-02845-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02845-6
This article is cited by
-
Pushing the limits of automated cell tracking
Nature Methods (2025)
-
Organoid: a promising solution to current challenges in cancer immunotherapy
npj Biomedical Innovations (2025)
-
Following a cell’s every move
Nature Methods (2025)