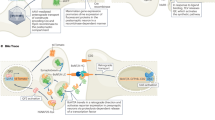
Abstract
Deciphering the connectome, the ensemble of synaptic connections that underlie brain function, is a central goal of neuroscience research. Here we report the in vivo mapping of connections between presynaptic and postsynaptic partners in zebrafish, by adapting the trans-Tango genetic approach that was first developed for anterograde transsynaptic tracing in Drosophila. Neural connections were visualized between synaptic partners in larval retina, brain and spinal cord and followed over development. The specificity of labeling was corroborated by functional experiments in which optogenetic activation of presynaptic spinal cord interneurons elicited responses in known motor neuronal postsynaptic targets, as measured by trans-Tango-dependent expression of a genetically encoded calcium indicator or by electrophysiology. Transsynaptic signaling through trans-Tango reveals synaptic connections in the zebrafish nervous system, providing a valuable in vivo tool to monitor and interrogate neural circuits over time.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The source data that support the findings of this study are available at Zenodo at https://doi.org/10.5281/zenodo.13716911 (ref. 85), https://doi.org/10.5281/zenodo.13629169 (ref. 89), https://doi.org/10.5281/zenodo.13629206 (ref. 90), https://doi.org/10.5281/zenodo.13629296 (ref. 91), https://doi.org/10.5281/zenodo.13629407 (ref. 92), https://doi.org/10.5281/zenodo.13629638 (ref. 93), https://doi.org/10.5281/zenodo.13629732 (ref. 94), https://doi.org/10.5281/zenodo.13629664 (ref. 95), https://doi.org/10.5281/zenodo.13629565 (ref. 96), https://doi.org/10.5281/zenodo.13629389 (ref. 97) and https://doi.org/10.5281/zenodo.13629333 (ref. 98). Additional raw imaging data are available upon request. We have the rights to publish BioRender figures and Fig. 1a was generated using BioRender. Source data are provided with this paper.
Code availability
The MATLAB and R codes used in the presented data analyses are publicly available at Zenodo at https://doi.org/10.5281/zenodo.13629206 (ref. 90). Modification to code for image registration84 is also provided.
Change history
10 November 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41593-025-02174-z
References
Glickstein, M. Golgi and Cajal: the neuron doctrine and the 100th anniversary of the 1906 Nobel Prize. Curr. Biol. 16, R147–R151 (2006).
Luo, L., Callaway, E. M. & Svoboda, K. Genetic dissection of neural circuits: a decade of progress. Neuron 98, 256–281 (2018).
Wickersham, I. R., Finke, S., Conzelmann, K. K. & Callaway, E. M. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat. Methods 4, 47–49 (2007).
Callaway, E. M. & Luo, L. Monosynaptic circuit tracing with glycoprotein-deleted rabies viruses. J. Neurosci. 35, 8979–8985 (2015).
Beier, K. T., Mundell, N. A., Pan, Y. A. & Cepko, C. L. Anterograde or retrograde transsynaptic circuit tracing in vertebrates with vesicular stomatitis virus vectors. Curr. Protoc. Neurosci. 74, 1.26.1–1.26.27 (2016).
Lichtman, J. W. & Denk, W. The big and the small: challenges of imaging the brain’s circuits. Science 334, 618–623 (2011).
Talay, M. et al. Transsynaptic mapping of second-order taste neurons in flies by trans-Tango. Neuron 96, 783–795 (2017).
Chen, Y. D., Park, S. J., Joseph, R. M., Ja, W. W. & Dahanukar, A. A. Combinatorial pharyngeal taste coding for feeding avoidance in adult Drosophila. Cell Rep. 29, 961–973 (2019).
Duhart, J. M., Baccini, V., Zhang, Y., Machado, D. R. & Koh, K. Modulation of sleep-courtship balance by nutritional status in. eLife 9, e60853 (2020).
Reinhard, N. et al. The neuronal circuit of the dorsal circadian clock neurons in. Front. Physiol. 13, 886432 (2022).
Sancer, G. et al. Modality-specific circuits for skylight orientation in the fly visual system. Curr. Biol. 29, 2812–2825 (2019).
Snell, N. J. et al. Complex representation of taste quality by second-order gustatory neurons in Drosophila. Curr. Biol. 32, 3758–3772 (2022).
Barnea, G. et al. The genetic design of signaling cascades to record receptor activation. Proc. Natl Acad. Sci. USA 105, 64–69 (2008).
Halpern, M. E. et al. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish 5, 97–110 (2008).
Subedi, A. et al. Adoption of the Q transcriptional regulatory system for zebrafish transgenesis. Methods 66, 433–440 (2014).
Kawakami, K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 8, S7 (2007).
Friedmann, D., Hoagland, A., Berlin, S. & Isacoff, E. Y. A spinal opsin controls early neural activity and drives a behavioral light response. Curr. Biol. 25, 69–74 (2015).
Park, H. C. et al. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev. Biol. 227, 279–293 (2000).
Xie, X. et al. Silencer-delimited transgenesis: NRSE/RE1 sequences promote neural-specific transgene expression in a NRSF/REST-dependent manner. BMC Biol. 10, 93 (2012).
Pisharath, H., Rhee, J. M., Swanson, M. A., Leach, S. D. & Parsons, M. J. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech. Dev. 124, 218–229 (2007).
Hoshino, M. et al. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron 47, 201–213 (2005).
Sternberg, J. R. et al. Optimization of a neurotoxin to investigate the contribution of excitatory interneurons to speed modulation in vivo. Curr. Biol. 26, 2319–2328 (2016).
Itoh, T. et al. Gsx2 is required for specification of neurons in the inferior olivary nuclei from Ptf1a-expressing neural progenitors in zebrafish. Development 147, dev190603 (2020).
Jusuf, P. R. & Harris, W. A. Ptf1a is expressed transiently in all types of amacrine cells in the embryonic zebrafish retina. Neural Dev. 4, 34 (2009).
Davison, C. & Zolessi, F. R. Slit2 is necessary for optic axon organization in the zebrafish ventral midline. Cells Dev. 166, 203677 (2021).
Ryu, S. et al. Orthopedia homeodomain protein is essential for diencephalic dopaminergic neuron development. Curr. Biol. 17, 873–880 (2007).
Pittman, A. J., Law, M. Y. & Chien, C. B. Pathfinding in a large vertebrate axon tract: isotypic interactions guide retinotectal axons at multiple choice points. Development 135, 2865–2871 (2008).
Tokumoto, M. et al. Molecular heterogeneity among primary motoneurons and within myotomes revealed by the differential mRNA expression of novel islet-1 homologs in embryonic zebrafish. Dev. Biol. 171, 578–589 (1995).
Thisse, B. et al. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 77, 505–519 (2004).
Knafo, S. et al. Mechanosensory neurons control the timing of spinal microcircuit selection during locomotion. eLife 6, e25260 (2017).
Daume, D., Offner, T., Hassenklöver, T. & Manzini, I. Patterns of tubb2b promoter-driven fluorescence in the forebrain of larval Xenopus laevis. Front. Neuroanat. 16, 914281 (2022).
Knafo, S. & Wyart, C. Active mechanosensory feedback during locomotion in the zebrafish spinal cord. Curr. Opin. Neurobiol. 52, 48–53 (2018).
Zheng, Y. Q. et al. Nexmifa regulates axon morphogenesis in motor neurons in zebrafish. Front. Mol. Neurosci. 15, 848257 (2022).
Sharrock, A. V. et al. NTR 2.0: a rationally engineered prodrug-converting enzyme with substantially enhanced efficacy for targeted cell ablation. Nat. Methods 19, 205–215 (2022).
Pisharath, H. & Parsons, M. J. Nitroreductase-mediated cell ablation in transgenic zebrafish embryos. Methods Mol. Biol. 546, 133–143 (2009).
Hoon, M., Okawa, H., Della Santina, L. & Wong, R. O. Functional architecture of the retina: development and disease. Prog. Retin Eye Res. 42, 44–84 (2014).
Schmitt, E. A. & Dowling, J. E. Comparison of topographical patterns of ganglion and photoreceptor cell differentiation in the retina of the zebrafish, Danio rerio. J. Comp. Neurol. 371, 222–234 (1996).
Nevin, L. M., Robles, E., Baier, H. & Scott, E. K. Focusing on optic tectum circuitry through the lens of genetics. BMC Biol. 8, 126 (2010).
Förster, D. et al. Retinotectal circuitry of larval zebrafish is adapted to detection and pursuit of prey. eLife 9, e58596 (2020).
Novales Flamarique, I. & Wachowiak, M. Functional segregation of retinal ganglion cell projections to the optic tectum of rainbow trout. J. Neurophysiol. 114, 2703–2717 (2015).
Scott, E. K. & Baier, H. The cellular architecture of the larval zebrafish tectum, as revealed by gal4 enhancer trap lines. Front. Neural Circuits 3, 13 (2009).
Sengupta, M., Daliparthi, V., Roussel, Y., Bui, T. V. & Bagnall, M. W. Spinal V1 neurons inhibit motor targets locally and sensory targets distally. Curr. Biol. 31, 3820–3833.e4 (2021).
Menelaou, E. & McLean, D. L. Hierarchical control of locomotion by distinct types of spinal V2a interneurons in zebrafish. Nat. Commun. 10, 4197 (2019).
Menelaou, E., VanDunk, C. & McLean, D. L. Differences in the morphology of spinal V2a neurons reflect their recruitment order during swimming in larval zebrafish. J. Comp. Neurol. 522, 1232–1248 (2014).
Bello-Rojas, S. & Bagnall, M. W. Clonally related, Notch-differentiated spinal neurons integrate into distinct circuits. eLife 11, e83680 (2022).
Higashijima, S., Masino, M. A., Mandel, G. & Fetcho, J. R. Engrailed-1 expression marks a primitive class of inhibitory spinal interneuron. J. Neurosci. 24, 5827–5839 (2004).
Freeman, J. et al. Mapping brain activity at scale with cluster computing. Nat. Methods 11, 941–950 (2014).
Wee, C. L. et al. A bidirectional network for appetite control in larval zebrafish. eLife 8, e43775 (2019).
Wang, W. C. & McLean, D. L. Selective responses to tonic descending commands by temporal summation in a spinal motor pool. Neuron 83, 708–721 (2014).
El Manira, A. Dynamics and plasticity of spinal locomotor circuits. Curr. Opin. Neurobiol. 29, 133–141 (2014).
Ahrens, M. B. et al. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature 485, 471–477 (2012).
Muto, A., Ohkura, M., Abe, G., Nakai, J. & Kawakami, K. Real-time visualization of neuronal activity during perception. Curr. Biol. 23, 307–311 (2013).
Antinucci, P. et al. A calibrated optogenetic toolbox of stable zebrafish opsin lines. eLife 9, e54937 (2020).
Ma, M., Kler, S. & Pan, Y. A. Structural neural connectivity analysis in zebrafish with restricted anterograde transneuronal viral labeling and quantitative brain mapping. Front. Neural Circuits 13, 85 (2019).
Satou, C. et al. A viral toolbox for conditional and transneuronal gene expression in zebrafish. eLife 11, e77153 (2022).
Asakawa, K. et al. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl Acad. Sci. USA 105, 1255–1260 (2008).
Bergeron, S. A. et al. Brain selective transgene expression in zebrafish using an NRSE derived motif. Front. Neural Circuits 6, 110 (2012).
Distel, M., Wullimann, M. F. & Köster, R. W. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc. Natl Acad. Sci. USA 106, 13365–13370 (2009).
Kawakami, K. et al. zTrap: zebrafish gene trap and enhancer trap database. BMC Dev. Biol. 10, 105 (2010).
Marquart, G. D. et al. A 3D searchable database of transgenic zebrafish Gal4 and Cre lines for functional neuroanatomy studies. Front. Neural Circuits 9, 78 (2015).
Akitake, C. M., Macurak, M., Halpern, M. E. & Goll, M. G. Transgenerational analysis of transcriptional silencing in zebrafish. Dev. Biol. 352, 191–201 (2011).
Goll, M. G., Anderson, R., Stainier, D. Y., Spradling, A. C. & Halpern, M. E. Transcriptional silencing and reactivation in transgenic zebrafish. Genetics 182, 747–755 (2009).
Pang, S. C., Wang, H. P., Zhu, Z. Y. & Sun, Y. H. Transcriptional activity and DNA methylation dynamics of the Gal4/UAS system in zebrafish. Mar. Biotechnol. 17, 593–603 (2015).
Baier, H. & Wullimann, M. F. Anatomy and function of retinorecipient arborization fields in zebrafish. J. Comp. Neurol. 529, 3454–3476 (2021).
Del Bene, F. & Wyart, C. Optogenetics: a new enlightenment age for zebrafish neurobiology. Dev. Neurobiol. 72, 404–414 (2012).
Robles, E., Smith, S. J. & Baier, H. Characterization of genetically targeted neuron types in the zebrafish optic tectum. Front. Neural Circuits 5, 1 (2011).
Sengupta, M. & Bagnall, M. W. Spinal interneurons: diversity and connectivity in motor control. Annu. Rev. Neurosci. 46, 79–99 (2023).
Lin, J. Y., Knutsen, P. M., Muller, A., Kleinfeld, D. & Tsien, R. Y. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat. Neurosci. 16, 1499–1508 (2013).
Ghosh, A. & Halpern, M. E. Transcriptional regulation using the Q system in transgenic zebrafish. Methods Cell Biol. 135, 205–218 (2016).
Burgess, J. et al. An optimized QF-binary expression system for use in zebrafish. Dev. Biol. 465, 144–156 (2020).
Fernandes, A. M. et al. Neural circuitry for stimulus selection in the zebrafish visual system. Neuron 109, 805–822 (2021).
Hong, J. et al. IQ-Switch is a QF-based innocuous, silencing-free, and inducible gene switch system in zebrafish. Commun. Biol. 4, 1405 (2021).
Sorkaç, A. et al. retro-Tango enables versatile retrograde circuit tracing in Drosophila. eLife 12, e85041 (2023).
Walker, C. Haploid screens and gamma-ray mutagenesis. Methods Cell Biol. 60, 43–70 (1999).
Parsons, M. J. et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech. Dev. 126, 898–912 (2009).
Fernandes, A. M. et al. Deep brain photoreceptors control light-seeking behavior in zebrafish larvae. Curr. Biol. 22, 2042–2047 (2012).
Ben Fredj, N. et al. Synaptic activity and activity-dependent competition regulates axon arbor maturation, growth arrest, and territory in the retinotectal projection. J. Neurosci. 30, 10939–10951 (2010).
Kimura, Y. et al. Hindbrain V2a neurons in the excitation of spinal locomotor circuits during zebrafish swimming. Curr. Biol. 23, 843–849 (2013).
Choi, J. H., Duboue, E. R., Macurak, M., Chanchu, J. M. & Halpern, M. E. Specialized neurons in the right habenula mediate response to aversive olfactory cues. eLife 10, e72345 (2021).
Kwan, K. M. et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 (2007).
Horstick, E. J. et al. Increased functional protein expression using nucleotide sequence features enriched in highly expressed genes in zebrafish. Nucleic Acids Res. 43, e48 (2015).
Suster, M. L., Abe, G., Schouw, A. & Kawakami, K. Transposon-mediated BAC transgenesis in zebrafish. Nat. Protoc. 6, 1998–2021 (2011).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Marquart, G. D. et al. High-precision registration between zebrafish brain atlases using symmetric diffeomorphic normalization. GigaScience 6, 1–15 (2017).
Coomer, C. Figure 3. Zenodo https://doi.org/10.5281/zenodo.13716911 (2024).
Drapeau, P., Ali, D. W., Buss, R. R. & Saint-Amant, L. In vivo recording from identifiable neurons of the locomotor network in the developing zebrafish. J. Neurosci. Methods 88, 1–13 (1999).
Masino, M. A. & Fetcho, J. R. Fictive swimming motor patterns in wild type and mutant larval zebrafish. J. Neurophysiol. 93, 3177–3188 (2005).
Heitler, W. J. Practical tools for analysing rhythmic neural activity. J. Neurosci. Methods 185, 151–164 (2009).
Coomer, C. Extended Fig. 5. Zenodo https://doi.org/10.5281/zenodo.13629169 (2024).
Coomer, C. Figure 6. Zenodo https://doi.org/10.5281/zenodo.13629206 (2024).
Coomer, C. Extended Fig. 3. Zenodo https://doi.org/10.5281/zenodo.13629296 (2024).
Coomer, C. Figure 2. Zenodo https://doi.org/10.5281/zenodo.13629407 (2024).
Coomer, C. Extended Fig. 1. Zenodo https://doi.org/10.5281/zenodo.13629638 (2024).
Coomer, C. Extended Fig. 4. Zenodo https://doi.org/10.5281/zenodo.13629732 (2024).
Coomer, C. Extended Fig. 2. Zenodo https://doi.org/10.5281/zenodo.13629664 (2024).
Coomer, C. Figure 4. Zenodo https://doi.org/10.5281/zenodo.13629565 (2024).
Coomer, C. Figure 5. Zenodo https://doi.org/10.5281/zenodo.13629389 (2024).
Coomer, C. Figure 1. Zenodo. https://doi.org/10.5281/zenodo.13629333 (2024).
Kani, S. et al. Proneural gene-linked neurogenesis in zebrafish cerebellum. Dev. Biol. 343, 1–17 (2010).
Acknowledgements
We thank M. Ahrens (HHMI, Janelia), M. Bagnall (Washington University), A. Douglas (The University of Utah), M. Granato (University of Pennsylvania), T. Kaminy (The University of Utah), K. Kwan (The University of Utah), J. Meserve (University of Pennsylvania), T. Mulligan (Johns Hopkins University) and J. Mumm (Johns Hopkins University) for sharing reagents and transgenic lines. We are grateful to E. Horstick for advice on codon optimization and to E. Spikol and A. Abdelfattah (Brown University) for expert guidance on calcium imaging experiments. Special thanks are extended to P. Robison for microscopy support, K. Young (Northwestern University) for technical assistance, and B. Malskis, J. Devine-Brilliant and C. Mathews for animal care. This work was supported by a Hanna H. Gray fellowship (GT15992 to C.E.C.), the National Institute of Neurological Disorders and Stroke (grants R21NS125187 and R21NS125207 to D.L.M.) and the National Institutes of Health Brain Initiative (RF1MH123213 to M.E.H., G.B., J.L. and D.R.).
Author information
Authors and Affiliations
Contributions
C.E.C., D.N., D.L.M. and M.E.H. conceived the experiments and wrote the manuscript with input from M.T., A.S., I.R., J.L., D.R. and G.B. C.E.C., D.N. and D.L.M. performed all experiments and data analysis. M.T., B.Z., N.J.S., A.S., J.M.C., J.C. and G.B. designed and generated trans-Tango plasmids. C.E.C., I.R., J.L. and D.R. advised on plasmids and collected preliminary data for zebrafish. J.C. applied image registration tools.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Claire Wyart and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Optimization of the trans-Tango ligand.
Six different ligand constructs were injected along with plasmids encoding the trans-Tango receptor, arrestin–TEV and Tol2 transposase RNA into single-cell zebrafish embryos, the progeny of Tg(QUAS:mApple-CAAX) and the Tg(ptf1a:Gal4-VP16) driver line that promotes strong expression of Tg(UAS:GFP) in the hindbrain. a–f, Ligands containing the transmembrane domain of zebrafish nrxn1a and varying lengths of mouse ICAM. a,a′, 10xUAS-E1B:sGCG-nrxn1a produced consistent trans-Tango labeling in the hindbrain (n = 20/25) but also nonspecific labeling (b; n = 15/25). c,c′, 10xUAS-E1B:sGCG-ICAM(1235)-nrxn1a produced consistent hindbrain labeling (n = 17/25) and nonspecific labeling (d; n = 11/25). e,e′, 10xUAS-E1B:sGCG-ICAM-nrxn1a produced fewer trans-Tango-labeled larvae (n = 6/25) and nonspecific labeling (f; n = 5/25). g–l, Ligands containing the transmembrane domain of zebrafish nrxn1b and varying lengths of mouse ICAM. g,g′, 10xUAS-E1B:sGCG-nrxn1b produced consistent trans-Tango labeling (n = 15/25) and substantial nonspecific labeling (h; n = 12/25). i,i′, 10xUAS-E1B:sGCG-ICAM(1235)-nrxn1b produced consistent labeling in the hindbrain (n = 23/25) and minimal nonspecific labeling (j; n = 2/25). k,k′, 10xUAS-E1B:sGCG-ICAM-nrxn1b produced trans-Tango labeling (n = 21/25) and nonspecific labeling (l; n = 14/25). Scale bars, 50 μm. Images are representative of trans-Tango labeling in the indicated number of larvae in five independent experiments for each ligand construct.
Extended Data Fig. 2 Monitoring synaptic connections over time.
a,a′, Dorsal view of mApple-CAAX-labeled neurons in the optic tectum (arrowheads) of a 6 dpf larva bearing Tg(isl2b.2:Gal4, myl7:tagRFP); Tg(UAS:GFP), Tg(QUAS:mApple-CAAX, he1.1:mCherry) and all trans-Tango components. Diffuse red fluorescent protein labeling (asterisk) is due to a secondary marker (my17:RFP) expressed in the heart. b,b′, In the same animal, labeling of the tectal neurons indicated in a (arrowheads) persisted at 2 weeks postfertilization (wpf; n = 10). Scale bars, 50 μm in a,a′ and 100 μm in b,b′. Images are representative of ten larvae from five independent experiments imaged at each time point.
Extended Data Fig. 3 Trans-Tango-mediated ablation of postsynaptic neurons.
a,b, In 5 dpf TgBAC(ptf1a:Gal4-VP16); Tg(UAS:GFP) larvae expressing the bacterial NfsB reductase gene, trans-Tango-labeled mApple-CAAX neurons (arrowheads) in merged (a) and single channel (b) images were no longer detected (c and d) following 1 day of incubation in 10 μM metronidazole (n = 25). c,d, Merged (c) and single channel (d) images. Scale bars, 50 μm. Images are representative of 25 larvae from 5 independent experiments imaged before (a and b) and after (c and d) metronidazole treatment.
Extended Data Fig. 4 Generation of trans-Tango transgenic zebrafish.
Heterozygous fish bearing transgenes for trans-Tango components and Tg(QUAS:mApple-CAAX, he1.1:mCherry) were mated with the TgBAC(pt1fa:Gal4-VP16)jh16 driver line and resultant embryos injected with plasmids for the other trans-Tango reagents and Tol2 RNA. a, RNA in situ hybridization indicates that the expression of UAS:sGCG-ICAM(1235)-Nrxn1b resembles the transcription pattern of the endogenous ptf1a gene99. b,c, Both the Tg(elavl3:hArr-TEV, he1.1:YFP)cd30 (b) and Tg(elavl:hGCGR-TEVcs-QF, he1.1:CFP) (c) stable lines are expressed broadly throughout the CNS. d,d″, Tg(10xUAS-E1B:sGCG-ICAM(1235)nrxn1b, cryaa:mCherry) larvae showed trans-Tango labeling in the hindbrain (n = 257/312) consistent with that observed when the ligand is introduced by plasmid injection. e,e″, Tg(elavl3:hArr-TEV, he1.1:YFP) larvae had similar trans-Tango labeling in the hindbrain (n = 132/164). f,f″, mApple-CAAX labeling was not observed in the hindbrain (n = 0/276) with the receptor line Tg(elavl:hGCGR-TEVcs-QF, he1.1:CFP) or when receptor plasmid was injected at a sub-threshold concentration (10 ng μl−1) along with the ligand and arrestin–TEV plasmids (25 ng µl−1; n = 25; g,g′). h,h′, However, the same concentration of receptor plasmid was effective at producing extensive mApple-CAAX labeling in the hindbrain (n = 21/25) when injected into embryos bearing Tg(elavl:hGCGR-TEVcs-QF, he1.1:CFP) along with the ligand and arrestin–TEV plasmids (25 ng µl−1). Scale bars, 20 µm for a–c and 50 μm for d–h. Images are representative of five independent experiments for each stable line.
Extended Data Fig. 5 Trans-Tango labeling in stable transgenic lines.
Robust mApple-CAAX labeling in Tg(isl2b.2:Gal4, myl7:tagRFP) 6 dpf larvae bearing both the ligand and arrestin–TEV stable transgenes and Tg(UAS:GFP) and Tg(QUAS:mApple-CAAX) reporter lines. a,a′, Control larvae from injections of plasmids for all trans-Tango components (n = 10). b,b′, Tg(10xUAS-sGCG-ICAM(1235)-nrxn1b, cryaa:mCherry) larvae injected with the receptor and arrestin–TEV constructs (n = 10). c,c′, Tg(elavl3:hArr:TEV, he1.1:YFP) larvae that had been injected with the receptor and ligand constructs (n = 10). d,d′, Larvae transgenic for both the ligand and arrestin–TEV transgenes that had been injected with the trans-Tango receptor construct (n = 10). e, Uninjected controls (n = 10). f, On average, 45 neurons were labeled with mApple-CAAX in larvae that bore both the ligand and arrestin–TEV stable transgenes compared to those from injections of all trans-Tango components (on average, 11 neurons per sample). In the boxplots the center line represents the median, the box limits indicate top and bottom quartiles, and the whiskers extend to the 1.5× quartile range. g, Image registration of mApple-CAAX-labeled neurons in the optic tectum and hindbrain of ten Tg(isl2b.2:Gal4-VP16, myl7:tagRFP); Tg(UAS:GFP); Tg(10xUAS-sGCG-ICAM(1235)-nrxn1b, cryaa:mCherry); Tg(elavl3:hArr:TEV, he1.1:YFP) larvae injected with the receptor construct. mApple-CAAX-labeled neurons are pseudocolored differently for each larva and dashed circles indicate some cells that were labeled in multiple samples. Scale bars, 20 μm. Double asterisk (**) represents the statistically significant increase (P < 0.0001) in trans-Tango labeled neurons when using the ligand and arrestin stable lines compared to plasmid only and stable lines individually using a one-way ANOVA test on Prism.
Supplementary information
Supplementary Tables
Supplementary Tables 1 and 2.
Source data
Source Data Extended Data Fig. 4
Statistical data from Prism used to calculate P value and statistical significance.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Coomer, C.E., Naumova, D., Talay, M. et al. Transsynaptic labeling and transcriptional control of zebrafish neural circuits. Nat Neurosci 28, 189–200 (2025). https://doi.org/10.1038/s41593-024-01815-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41593-024-01815-z