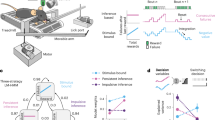
Abstract
The cognitive processes supporting complex animal behavior are closely associated with movements responsible for critical processes, such as facial expressions or the active sampling of our environments. These movements are strongly related to neural activity across much of the brain and are often highly correlated with ongoing cognitive processes. A fundamental issue for understanding the neural signatures of cognition and movements is whether cognitive processes are separable from related movements or if they are driven by common neural mechanisms. Here we demonstrate how the separability of cognitive and motor processes can be assessed and, when separable, how the neural dynamics associated with each component can be isolated. We designed a behavioral task in mice that involves multiple cognitive processes, and we show that dynamics commonly taken to support cognitive processes are strongly contaminated by movements. When cognitive and motor components are isolated using a novel approach for subspace decomposition, we find that they exhibit distinct dynamical trajectories and are encoded by largely separate populations of cells. Accurately isolating dynamics associated with particular cognitive and motor processes will be essential for developing conceptual and computational models of neural circuit function.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The data described in this study are available on Zenodo at https://doi.org/10.5281/zenodo.13941414 (ref. 69).
Code availability
MATLAB and Python code for subspace identification is available at https://github.com/economolab/subspaceID. Custom MATLAB code used for analyses is available on Zenodo at https://doi.org/10.5281/zenodo.13941414 (ref. 69).
References
Alexander, G. E. & Crutcher, M. D. Neural representations of the target (goal) of visually guided arm movements in three motor areas of the monkey. J. Neurophysiol. 64, 164–178 (1990).
Cisek, P. & Kalaska, J. F. Neural correlates of reaching decisions in dorsal premotor cortex: specification of multiple direction choices and final selection of action. Neuron 45, 801–814 (2005).
Chen, S. et al. Brain-wide neural activity underlying memory-guided movement. Cell 187, 676–691 (2024).
Zimnik, A. J. & Churchland, M. M. Independent generation of sequence elements by motor cortex. Nat. Neurosci. 24, 412–424 (2021).
Ames, K. C., Ryu, S. I. & Shenoy, K. V. Simultaneous motor preparation and execution in a last-moment reach correction task. Nat. Commun. 10, 2718 (2019).
Fuster, J. M. & Alexander, G. E. Neuron activity related to short-term memory. Science 173, 652–654 (1971).
Erlich, J. C., Bialek, M. & Brody, C. D. A cortical substrate for memory-guided orienting in the rat. Neuron 72, 330–343 (2011).
Tanji, J. & Evarts, E. V. Anticipatory activity of motor cortex neurons in relation to direction of an intended movement. J. Neurophysiol. 39, 1062–1068 (1976).
Guo, Z. V. et al. Flow of cortical activity underlying a tactile decision in mice. Neuron 81, 179–194 (2014).
Wallis, J. D., Anderson, K. C. & Miller, E. K. Single neurons in prefrontal cortex encode abstract rules. Nature 411, 953–956 (2001).
Schultz, W., Dayan, P. & Montague, P. R. A neural substrate of prediction and reward. Science 275, 1593–1599 (1997).
Freedman, D. J., Riesenhuber, M., Poggio, T. & Miller, E. K. Categorical representation of visual stimuli in the primate prefrontal cortex. Science 291, 312–316 (2001).
Zagha, E. The importance of accounting for movement when relating neuronal activity to sensory and cognitive processes. J. Neurosci. 42, 1375–1382 (2022).
Drew, P. J., Winder, A. T. & Zhang, Q. Twitches, blinks, and fidgets: important generators of ongoing neural activity. Neuroscientist 25, 298–313 (2019).
Hulsey, D., Zumwalt, K., Mazzucato, L., McCormick, D. A. & Jaramillo, S. Decision-making dynamics are predicted by arousal and uninstructed movements. Cell Rep. 43, 113709 (2024).
Tremblay, S., Testard, C., DiTullio, R. W., Inchauspé, J. & Petrides, M. Neural cognitive signals during spontaneous movements in the macaque. Nat. Neurosci. 26, 295–305 (2023).
Musall, S., Kaufman, M. T., Juavinett, A. L., Gluf, S. & Churchland, A. K. Single-trial neural dynamics are dominated by richly varied movements. Nat. Neurosci. 22, 1677–1686 (2019).
Stringer, C. Spontaneous behaviors drive multidimensional, brainwide activity. Science 364, 255 (2019).
Steinmetz, N. A., Zatka-Haas, P., Carandini, M. & Harris, K. D. Distributed coding of choice, action and engagement across the mouse brain. Nature 576, 266–273 (2019).
Mangin, E. N., Chen, J., Lin, J. & Li, N. Behavioral measurements of motor readiness in mice. Curr. Biol. 33, 3610–3624 (2023).
Terada, S.-I., Kobayashi, K. & Matsuzaki, M. Transition of distinct context-dependent ensembles from secondary to primary motor cortex in skilled motor performance. Cell Rep. 41, 111494 (2022).
Lowet, E. et al. Enhanced neural processing by covert attention only during microsaccades directed toward the attended stimulus. Neuron 99, 207–214 (2018).
Popescu, S. T. & Wexler, M. Spontaneous body movements in spatial cognition. Front. Psychol. 3, 136 (2012).
Hajnal, J. V. et al. Artifacts due to stimulus correlated motion in functional imaging of the brain. Magn. Reson. Med. 31, 283–291 (1994).
Kaufman, M. T., Churchland, M. M., Ryu, S. I. & Shenoy, K. V. Cortical activity in the null space: permitting preparation without movement. Nat. Neurosci. 17, 440–448 (2014).
Cisek, P. & Kalaska, J. F. Neural mechanisms for interacting with a world full of action choices. Annu. Rev. Neurosci. 33, 269–298 (2010).
Elsayed, G. F., Lara, A. H., Kaufman, M. T., Churchland, M. M. & Cunningham, J. P. Reorganization between preparatory and movement population responses in motor cortex. Nat. Commun. 7, 13239 (2016).
Druckmann, S. & Chklovskii, D. B. Neuronal circuits underlying persistent representations despite time varying activity. Curr. Biol. 22, 2095–2103 (2012).
Galiñanes, G. L., Bonardi, C. & Huber, D. Directional reaching for water as a cortex-dependent behavioral framework for mice. Cell Rep. 22, 2767–2783 (2018).
Mathis, A. et al. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281–1289 (2018).
Mayrhofer, J. M. et al. Distinct contributions of whisker sensory cortex and tongue-jaw motor cortex in a goal-directed sensorimotor transformation. Neuron 103, 1034–1043 (2019).
Inagaki, H. K. et al. Neural algorithms and circuits for motor planning. Annu. Rev. Neurosci. 45, 249–271 (2022).
Xu, D. et al. Cortical processing of flexible and context-dependent sensorimotor sequences. Nature 603, 464–469 (2022).
Yin, X., Wang, Y., Li, J. & Guo, Z. V. Lateralization of short-term memory in the frontal cortex. Cell Rep. 40, 111190 (2022).
Li, N., Chen, T.-W., Guo, Z. V., Gerfen, C. R. & Svoboda, K. A motor cortex circuit for motor planning and movement. Nature 519, 51–56 (2015).
Chen, T.-W., Li, N., Daie, K. & Svoboda, K. A map of anticipatory activity in mouse motor cortex. Neuron 94, 866–879 (2017).
Shenoy, K. V., Sahani, M. & Churchland, M. M. Cortical control of arm movements: a dynamical systems perspective. Annu. Rev. Neurosci. 36, 337–359 (2013).
Li, N., Daie, K., Svoboda, K. & Druckmann, S. Robust neuronal dynamics in premotor cortex during motor planning. Nature 532, 459–464 (2016).
Yang, W., Tipparaju, S. L., Chen, G. & Li, N. Thalamus-driven functional populations in frontal cortex support decision-making. Nat. Neurosci. 25, 1339–1352 (2022).
Economo, M. N. et al. Distinct descending motor cortex pathways and their roles in movement. Nature 563, 79–84 (2018).
Narayanan, N. S. Ramping activity is a cortical mechanism of temporal control of action. Curr. Opin. Behav. Sci. 8, 226–230 (2016).
Cisek, P., Puskas, G. A. & El-Murr, S. Decisions in changing conditions: the urgency-gating model. J. Neurosci. 29, 11560–11571 (2009).
Wang, Z. A. et al. Not everything, not everywhere, not all at once: a study of brain-wide encoding of movement. Preprint at bioRxiv https://doi.org/10.1101/2023.06.08.544257 (2023).
Inagaki, H. K., Fontolan, L., Romani, S. & Svoboda, K. Discrete attractor dynamics underlies persistent activity in the frontal cortex. Nature 566, 212–217 (2019).
Moore, J. D., Kleinfeld, D. & Wang, F. How the brainstem controls orofacial behaviors comprised of rhythmic actions. Trends Neurosci. 37, 370–380 (2014).
Arber, S. & Costa, R. M. Networking brainstem and basal ganglia circuits for movement. Nat. Rev. Neurosci. 23, 342–360 (2022).
Svoboda, K. & Li, N. Neural mechanisms of movement planning: motor cortex and beyond. Curr. Opin. Neurobiol. 49, 33–41 (2018).
Christensen, A. J., Ott, T. & Kepecs, A. Cognition and the single neuron: how cell types construct the dynamic computations of frontal cortex. Curr. Opin. Neurobiol. 77, 102630 (2022).
Gallego, J. A., Perich, M. G., Miller, L. E. & Solla, S. A. Neural manifolds for the control of movement. Neuron 94, 978–984 (2017).
Dahmen, D. et al. Strong and localized recurrence controls dimensionality of neural activity across brain areas. Preprint at bioRxiv https://doi.org/10.1101/2020.11.02.365072 (2023).
Morales-Gregorio, A. et al. Neural manifolds in V1 change with top-down signals from V4 targeting the foveal region. Cell Rep. 43, 114371 (2024).
Mante, V., Sussillo, D., Shenoy, K. V. & Newsome, W. T. Context-dependent computation by recurrent dynamics in prefrontal cortex. Nature 503, 78–84 (2013).
Joshi, S. & Gold, J. I. Pupil size as a window on neural substrates of cognition. Trends Cogn. Sci. 24, 466–480 (2020).
Pereira, T. D., Shaevitz, J. W. & Murthy, M. Quantifying behavior to understand the brain. Nat. Neurosci. 23, 1537–1549 (2020).
Miller, C. T. et al. Natural behavior is the language of the brain. Curr. Biol. 32, R482–R493 (2022).
Datta, S. R., Anderson, D. J., Branson, K., Perona, P. & Leifer, A. Computational neuroethology: a call to action. Neuron 104, 11–24 (2019).
Dennis, E. J. et al. Systems neuroscience of natural behaviors in rodents. J. Neurosci. 41, 911–919 (2021).
Kim, T. H. et al. Long-term optical access to an estimated one million neurons in the live mouse cortex. Cell Rep. 17, 3385–3394 (2016).
Jun, J. J. et al. Fully integrated silicon probes for high-density recording of neural activity. Nature 551, 232–236 (2017).
Jun, J. J. et al. Real-time spike sorting platform for high-density extracellular probes with ground-truth validation and drift correction. Preprint at bioRxiv https://doi.org/10.1101/101030 (2017).
Pachitariu, M., Sridhar, S., Pennington, J. & Stringer, C. Spike sorting with Kilosort4. Nat. Methods 21, 914–921 (2024).
Vincent, J. P. & Economo, M. N. Assessing cross-contamination in spike-sorted electrophysiology data. eNeuro https://doi.org/10.1523/ENEURO.0554-23.2024 (2024).
Altan, E., Solla, S. A., Miller, L. E. & Perreault, E. J. Estimating the dimensionality of the manifold underlying multi-electrode neural recordings. PLoS Comput. Biol. 17, e1008591 (2021).
Horn, J. L. A rationale and test for the number of factors in factor analysis. Psychometrika 30, 179–185 (1965).
Boumal, N., Mishra, B., Absil, P.-A. & Sepulchre, R. Manopt, a Matlab toolbox for optimization on manifolds. J. Mach. Learn. Res. 15, 1455–1459 (2014).
Jiang, X., Saggar, H., Ryu, S. I., Shenoy, K. V. & Kao, J. C. Structure in neural activity during observed and executed movements is shared at the neural population level, not in single neurons. Cell Rep. 32, 108006 (2020).
Aarts, E., Verhage, M., Veenvliet, J. V., Dolan, C. V. & van der Sluis, S. A solution to dependency: using multilevel analysis to accommodate nested data. Nat. Neurosci. 17, 491–496 (2014).
Leeden, R.v., Meijer, E. & Busing, F. M. in Handbook of Multilevel Analysis (eds Leeuw, J. & Meijer, E.) 401–433 (Springer, 2008).
Hasnain, M., Birnbaum, J. & Economo, M. Data and code accompanying Hasnain, Birnbaum et al., Nature Neuroscience 2024 [data set]. Zenodo https://doi.org/10.5281/zenodo.13941415 (2024).
Acknowledgements
We thank K. Svoboda, B. DePasquale, S. Druckmann and B. Scott for helpful discussions; T. Wang for helpful comments on the manuscript; and J. Jiang for help with crystal skull surgeries. This work was supported by the Whitehall Foundation, the Klingenstein Fund, the Simons Foundation and National Institutes of Health R01NS121409 and U19NS137920.
Author information
Authors and Affiliations
Contributions
M.A.H., J.E.B. and M.N.E. conceived of the project. M.N.E. and C.C. supervised research. M.A.H., J.E.B. and M.N.E. designed experiments. M.A.H., J.E.B., J.L.U.N. and E.K.H. performed experiments. M.A.H., J.E.B. and M.N.E. analyzed data. M.A.H., J.E.B. and M.N.E. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Andrei Khilkevich, Thomas Mrsic-Flogel and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Region-specific photoinactivation of ALM and tjM1.
a. Percentage of time spent moving (see Methods) in a session determined by video recordings using the side view, bottom view, and both views. Points indicate each of the 12 randomly selected sessions used for this analysis. Error bars denote standard deviation across sessions (n = 12). b. Effect of delay epoch photoinactivation on behavioral performance (middle column) and uninstructed movements (right column) when photoinactivation was directed to the MC (ALM + tjM1; top row; same data as Fig. 1g–in = 14 sessions, 2 mice), the ALM (middle row, n = 15 sessions, 2 mice), and the tjM1 (bottom, n = 9 sessions, 2 mice). Photoinactivation of ALM and tjM1 led to similar behavioral impairment and reduction in uninstructed movements, with larger effects observed with MC (ALM + tjM1) photoinactivation. c. Tongue length during control and go cue/water drop photoinactivation trials for delayed-response (left) and water-cued (right) contexts. Blue traces indicate right lickport contacts, red traces indicate left contacts, and black traces indicate no contact. Vertical dashed line indicates go cue or water drop onset. Blue shaded region indicates photoinactivation period. d. Percentage of time with tongue visible during photoinactivation period for DR trials (left) and WC trials (right). Each colored point indicates mean value for an animal (n = 4 animals), individual animals are connected by black lines. Light gray lines denote individual sessions (n = 10 sessions). Bars are the mean across all sessions. Asterisks denote significant differences (p < 0.05) between control and photoinactivation trials (Percent reduction on all DR trials: 19 ± 7%, mean ± s.d., p = 1.6e-05; DR left trials: 20 ± 8%, p = 3.0e-05; DR right trials: 20 ± 7%, p = 1.6e-05; All WC trials: 4 ± 8%, p = 0.154; WC left trials: 1% ± 9%, p = 0.702; WC right trials: 7% ± 5%, p = 0.002; paired two-sided t-test, n = 10 sessions). Error bars indicate standard deviation across sessions. In WC trials, tongue protrusion was only significantly impaired on one trial type, while ability to successfully contact the lickport was impaired in all conditions (see Fig. 1c).
Extended Data Fig. 2 Session-by-session statistics.
a. Number of recorded single- and multi-units per session for the fixed delay task (left) or the randomized delay task (right). Left, Purple shaded region indicates sessions in which animals were only presented with DR trials. Green and blue bars underneath plots indicate the probe type used for a given session. b. Variance explained of trial-averaged neural activity by each coding direction. The coding directions were calculated using neural activity from individual sessions (n = 25). Bar height represents the mean across sessions and error bars indicate standard deviation across sessions. c. Receiver operating curves (ROC) demonstrating choice decoding accuracy from delay epoch CDchoice projections across all individual sessions (see Methods). Inset: area under the ROC curve (AUC). Bar height represents the mean across sessions and points indicate sessions.
Extended Data Fig. 3 Session-by-session variability in the relationship between kinematic features and putative cognitive dynamics.
a. Regression weights for each group of kinematic predictors of CDchoice projections, as a fraction of all predictor coefficients (see Methods). Sessions are sorted in descending order by motion energy fraction. Outlined bars indicate example sessions shown in (b) and (c). b. Example session where motion energy made up the largest fraction of regression weights for predicting CDchoice projections. Top, overlayed jaw/nose speed or motion energy for a subset trials. Bottom, two example trials of kinematic feature trajectories. c. Same as (b) but for an example session where jaw and nose features made up a larger fraction of regression weights. d. Same as (a) but for predicting CDcontext projections. e, f. Same as (b) and (c) but for two example sessions with different regression weight fractions for predicting CDcontext projections.
Extended Data Fig. 4 Control analyses for subspace decomposition.
a, b. Movement-null and movement-potent subspaces estimated as in Fig. 5g–l using DR and WC trials. a. Variance explained (R2) of motion energy by the sum squared magnitude of activity in the movement-null and movement-potent subspaces on single trials. Each point is the mean across trials for a session. b. Left, motion energy on single trials for an example session. Middle, sum-squared magnitude of activity in the movement-potent subspace. Right, sum-squared magnitude of activity in the movement-null subspace. Trials sorted by average delay epoch motion energy. c. Selectivity (left vs. right) of the neural population during WC trials. Mean and 95% CI across sessions shown. d, e. Same as (a,b) but estimating movement-null and movement-potent subspaces using WC trials only. f. Normalized magnitude of activity in the movement-null subspace (left) movement-potent subspace (right) when estimated using DR and WC trials as in (a,b), versus when estimated using WC trials only as in (d,e). Circles are average activity per trial for an example session. g, h. Same as (a,b), but estimating movement-null and movement-potent subspaces using data restricted to the response epoch of DR and WC trials. i. Magnitude of activity in the movement-null subspace (left) or movement-potent subspace (right) when estimated using DR and WC trials as in (a,b) versus when estimated using data from only the response epoch of DR and WC trials as in (g,h). Circles are average activity per trial for an example session. j, k. Same as (a,b), but estimating the movement-null and movement-potent subspaces using a two-stage PCA approach (see Methods). This approach is conservative in avoiding the mis-assignment of cognitive dynamics that correlate in time with movement to the movement-potent subspace. l. Magnitude of activity in the movement-null subspace (left) or movement-potent subspace (right) when estimated using DR and WC trials as in (a,b) versus when estimated using the two-stage PCA approach as in (j,k). Circles are average activity per trial for an example session.
Extended Data Fig. 5 Alignment of single-units to random subspaces.
Random subspaces were constructed by independently and identically drawing from a normal distribution with zero mean and unit variance. Each random subspace was then biased towards the covariance structure of the actual data (see Methods). a. Null distributions of alignment indices for trial-averaged data. b. Null distributions of alignment indices for single-trial data. Null alignment distributions are skewed towards the movement-potent subspace due to the unbalanced variance between delay and response epochs (a) or between stationary and movement time points (b), reflecting the strong movement tuning of many units.
Extended Data Fig. 6 Varying dimensionality of subspaces.
Analyses were repeated while varying the dimensionality of movement-null and movement-potent subspaces. Each subspace was constrained to be 4 (left), 6 (middle left), 8 (middle right), or 13 dimensions (right). a. Upper bound estimate of dimensionality for trial-averaged (PSTH) data and single-trial data. Bar heights indicate mean across sessions, points indicate sessions, and error bars indicate standard deviation across sessions (n = 25 sessions). b. Cumulative variance explained of the neural activity by the activity in movement-null and movement-potent subspaces. Bold lines and points indicate mean across sessions. Thin lines represent single sessions c. Normalized variance explained of neural activity during the delay or response epoch by the activity in movement-null and movement-potent subspaces. Points indicate sessions, bar height indicates mean across sessions, and error bars indicate standard deviation across sessions (n = 25 sessions). d–f. Subspace (d), CDchoice (e), and CDramp (f) alignment distributions when varying dimensionalities of each subspace.
Extended Data Fig. 7 Projections along movement-null and movement-potent components of CDchoice.
a. Same data as in Fig. 7a except all time in trial shown to highlight activity during the response epoch. Selectivity (projections onto CDchoice on lick-right trials minus projections on lick-left trials) of movement-null (left) and movement-potent (right) subspace activity. Mean and 5–95% CI of the bootstrap distribution for correct (solid) and error (dashed) trials shown. b. Change in selectivity between the last 100 ms of the delay epoch and the last 100 ms of the sample epoch in movement-null and movement-potent components of projections along CDchoice (Movement-potent: 2.25 ± 1.57, mean ± s.d., Movement-null: 0.76 ± 0.8, p = 1 × 10−5, paired two-sided t-test, n = 25 sessions). Points indicate individual sessions, bar height indicates mean across sessions, and error bars indicate standard deviation across sessions. c. Three example sessions from three different mice depicting selectivity along CDchoice as in Fig. 7a. Solid lines denote the mean projection on correct trials and dashed lines denote the mean projection on error trials.
Extended Data Fig. 8 Within-subspace CD projections using variations on procedure to determine subspaces.
a. Projections of movement-null and movement-potent subspace activity along CDramp for each of three analytical variations. Movement-null and movement-potent subspaces were identified using both DR and WC trials (left), WC trials only (middle), and the response epoch of DR and WC trials (right). Mean and 95% CI of bootstrap distribution shown. b. Projections along movement-null (left) and movement-potent (right) components of CDramp when determined from activity within each subspace individually, rather than from the full neural population. Mean and 95% CI of bootstrap distribution shown.
Extended Data Fig. 9 Encoding of context in both the null and potent subspaces tracks block-wise task structure.
a. Heatmap of single-trial projections of null and potent subspace activity along CDcontext for an example session. The chronological DR or WC block within the session is denoted by differently shaded purple and orange rectangles, respectively, on the right of each plot. b. Same as (a) but for another example session, from a different animal.
Extended Data Fig. 10 Relationship between tongue angle and neural activity in the movement-null and movement-potent subspaces.
a. Projections along movement-potent (top) and movement-null (bottom) components of CDaction. Correct trials shown in solid lines and error trials shown in dashed lines. Shaded region depicts 95% CI of bootstrap distribution. b. Tongue angle for an example session for correct and error trials Black values indicate tongue not visible. c. Tongue angle on correct and error right and left trials. Tongue angle was linearly time warped to allow for averaging over trials and sessions. Mean and s.e.m. across sessions shown. d. Tongue angle (left) and predictions from the full population neural activity (middle left), null subspace activity (middle right), and potent subspace activity (right) for an example session. e. Variance explained (R2) of tongue angle by prediction from movement-null (green) and movement-potent (pink) subspaces. Asterisks denote significant differences between predictions from null and potent subspaces and error bars indicate standard deviation across sessions (p = 2 × 10−8, paired two-sided t-test, n = 25 sessions).
Supplementary information
Supplementary Information
Supplementary Notes 1 and 2.
Supplementary Movie 1
Uninstructed movements vary in their identity and timing. Example trials in which uninstructed movements vary in their identity (across rows) and timing (across columns). Traces represent the y position of the feature within the video frame. All example trials are taken from the same mouse and session.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hasnain, M.A., Birnbaum, J.E., Ugarte Nunez, J.L. et al. Separating cognitive and motor processes in the behaving mouse. Nat Neurosci 28, 640–653 (2025). https://doi.org/10.1038/s41593-024-01859-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41593-024-01859-1
This article is cited by
-
How distributed is the brain-wide network that is recruited for cognition?
Nature Reviews Neuroscience (2026)
-
Brain-wide analysis reveals movement encoding structured across and within brain areas
Nature Neuroscience (2026)
-
Facial expressions in mice reveal latent cognitive variables and their neural correlates
Nature Neuroscience (2025)