Abstract
Tumor suppressor p53-binding protein 1 (53BP1) regulates DNA end joining in lymphocytes, diversifying immune antigen receptors. This involves nucleosome-bound 53BP1 at DNA double-stranded breaks (DSBs) recruiting Rap1-interacting factor 1 homolog (RIF1) and shieldin, a poorly understood DNA-binding complex. The 53BP1–RIF1–shieldin axis is pathological in BRCA1-mutated cancers, blocking homologous recombination (HR) and driving illegitimate nonhomologous end joining (NHEJ). However, how this axis regulates DNA end joining and HR suppression remains unresolved. We investigated shieldin and its interplay with the Ctc1–Stn1–Ten1 (CST) complex, which was recently implicated downstream of 53BP1. Immunophenotypically, mice lacking shieldin or CST are equivalent, with class-switch recombination coreliant on both complexes. Ataxia-telangiectasia mutated kinase-dependent DNA damage signaling underpins this cooperation, inducing physical interactions between these complexes that reveal shieldin as a DSB-responsive CST adaptor. Furthermore, DNA polymerase ζ functions downstream of shieldin, establishing DNA fill-in synthesis as the physiological function of shieldin–CST. Lastly, we demonstrate that 53BP1 suppresses HR and promotes NHEJ in BRCA1-deficient mice and cells independently of shieldin. These findings showcase the versatility of the 53BP1 pathway, achieved through the collaboration of chromatin-bound 53BP1 complexes and DNA end-processing effector proteins.
Similar content being viewed by others
Main
DNA double-strand breaks (DSBs) are highly toxic and must be repaired accurately to counteract the threat of human disease and oncogenic mutations1. Paradoxically, mutagenic DSB repair by nonhomologous end joining (NHEJ) is favored in developing and antigen-stimulated lymphocytes, where it mediates deletional recombination events that diversify the antigenic specificity and function of B and T cell receptors2. To cope with this intrinsic discrepancy in desired DNA repair outcome between different cellular contexts, cells have evolved complex regulatory systems that maintain an appropriate equilibrium between competing DNA repair pathways, ensuring that DNA breaks are appropriately resolved3.
The generation of functional B and T cell receptors by the mammalian adaptive immune system relies on two programmed gene rearrangement mechanisms: V(D)J recombination and class-switch recombination (CSR)2. Both mechanisms rely on intragenic deletional recombination events, which are mediated through the ligation of interspaced DNA ends using NHEJ pathways. Because of the distinct biochemical mechanisms involved in generating DSBs during V(D)J recombination and CSR, the resulting DNA ends have different structural properties, necessitating distinct DNA end-processing and ligation mechanisms4.
DSB induction during V(D)J recombination occurs during the development of B and T cells in the bone marrow and thymus, respectively. The process is orchestrated by recombination-activating gene enzymes 1 and 2 (RAG1/2), which cleave V, D and J segment-flanking recombination signal sequences (RSSs), resulting in pairs of blunt signal and covalently sealed hair-pinned coding ends. Hairpin opening is essential before coding joins can occur, a step facilitated by the Artemis endonuclease upon its activation by the DNA-dependent protein kinase (DNA-PK) complex. This generates DNA ends that are strictly joined by classical NHEJ (c-NHEJ) proteins including Ku (Ku70 and Ku80), X-ray repair cross-complementing protein 4 (XRCC4) and DNA ligase 4 (LIG4), with a more modest reliance on XRCC4-like factor (XLF)2.
In contrast, CSR in mature peripheral lymphocytes involves DSB generation between antibody isotype-encoding constant (C) gene segments of the immunoglobulin heavy chain locus (igh). This process is initiated by activation-induced cytidine deaminase (AID), which introduces clusters of U•G mismatches in actively transcribed switch (S) introns. These mismatches are then processed by base excision repair (BER) and mismatch repair (MMR) machineries, ultimately leading to the conversion of single-stranded DNA (ssDNA) gaps into DSBs with single-stranded tailed termini. Unlike RAG1/2-induced DSBs, AID-dependent DSBs during CSR can be repaired through both c-NHEJ and alternative end joining (a-EJ) pathways, with the tumor suppressor p53-binding protein 1 (53BP1) pathway proteins also having a critical role4.
The 53BP1 pathway is composed of nucleosomal and DNA-binding protein components. The most upstream component is 53BP1, a chromatin reader protein scaffold that interacts with the DSB-responsive ubiquitinated Lys15 on nucleosomal H2A type histones (H2AK15ub) and ubiquitous histone H4 methylation at Lys20 (H4K20me1/2)5,6. This interaction leads to the formation of extensive 53BP1 chromatin-bound domains at DSB sites. Downstream of 53BP1, the 53BP1-binding protein Rap1-interacting factor 1 homolog (RIF1) bridges interactions between Ataxia-telangiectasia mutated kinase (ATM)-phosphorylated motifs in 53BP1 and the SHLD3 subunit of shieldin7,8, a complex bound to ssDNA that is thought to protect DNA ends9. Studies using genetically engineered mouse models deleted of 53bp1 or shieldin proteins Rev7 or Shld1/2 present with near-complete deficiencies in CSR. By contrast, only 53bp1−/− mice exhibit V(D)J recombination defects, which lead to mild B and T cell lymphopenia10,11,12,13.
The importance of the 53BP1 pathway extends to BRCA1-mutated cancers. In normal cells, the BRCA1 (breast cancer type 1 susceptibility protein)–BARD1 (BRCA1-associated RING domain 1) heterodimer binds to a related combinatorial chromatin signature comprising H2AK15ub and unmethylated Lys20 on histone H4 (a postreplicative modification state), excluding 53BP1 from DNA damage sites and directing homologous recombination (HR)14,15. However, in BRCA1-deficient cells, uninhibited 53BP1–chromatin complexes prevent HR and promote NHEJ-mediated genome rearrangements, leading to genomic instability16,17. Consequently, BRCA1-deficient cells and tumors are hypersensitive to poly(ADP-ribose) polymerase (Pol) inhibitors (PARPis), which exploit the HR defect caused by 53BP1 (ref. 16). Conversely, 53BP1 deletion is able to restore PARPi resistance to BRCA1-mutant cells18 and can even rescue viability in Brca1-null mice19,20.
Genetic screens aimed at elucidating PARPi resistance mechanisms in BRCA1-deficient cells were instrumental in revealing the 53BP1 pathway biology. Shieldin genes were discovered through screens investigating PARPi-induced toxicity in BRCA1-deficient cells21,22,23 and the Pol α–primase accessory complex proteins Ctc1–Stn1–Ten1 (CST) similarly emerged as putative 53BP1 pathway effectors24. Concurrent hypothesis-guided studies also linked the activities of shieldin and CST to pathological 53BP1 pathway function in BRCA1-mutant cells11,25,26,27,28 and highlighted CST-directed Pol α–primase-dependent DNA fill-in synthesis as a key intermediary step27,29,30.
This research has ignited debates regarding the precise role of shieldin, particularly whether its primary function involves blocking nucleolytic resection or facilitating fill-in synthesis at DNA ends4,31. The importance of effector interactions in the 53BP1 pathway during physiological antigen receptor recombination mechanisms, as well as their impact on HR defects and DNA joining events in BRCA1-deficient cells, remains to be fully understood. To address these questions, we conducted a comprehensive analysis using genetically engineered mouse models. Our findings demonstrate that the 53BP1 pathway contributes substantially to DNA end joining during antigen receptor recombination and in cancer cells in the absence of shieldin and CST. We also demonstrate a strict interdependence between shieldin and CST during CSR and an epistatic relationship with DNA Pol ζ, solidifying fill-in synthesis as a primary shieldin-dependent DNA repair function. Importantly, using BRCA1-deficient mice and cellular models, we establish that HR suppression relies on 53BP1 but not shieldin. Altogether, our results showcase remarkable resilience in the 53BP1 pathway, achieved through a division of labor between chromatin-bound 53BP1 complexes and their downstream DNA-bound effectors.
Results
Shieldin and CST equivalency during lymphocyte development
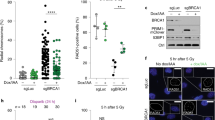
To understand the extent to which shieldin–CST cooperation supports physiological 53BP1 pathway function, we generated new mouse strains deleted for genes encoding core proteins in each complex in an inbred C57BL/6 background. Knockout mice deleted for critical exons 4–5 in Shld2 or exon 2 in Shld3, which comprises its entire coding sequences (hereafter referred to as Shld2−/− and Shld3−/− mice; Fig. 1a and Extended Data Fig. 1a,b), were viable, healthy, fertile and born at expected Mendelian frequencies. Moreover, genomic instability in 8–12-week-old Shld2−/− and Shld3−/− mice did not exceed that seen in aged-matched cohorts of wild-type (WT) or 53bp1−/− mice, as defined by levels of micronuclei in erythrocytes (Extended Data Fig. 1d,e), a proxy for systemic genomic instability32.
a, Schematic representation of the Shld2 and Shld3 (knockout) and Ctc1 (conditional) alleles generated in this study. TSS, transcription start site. b, Absolute numbers of B220+ B cells in the bone marrow (one femur and one tibia) and spleen (n = 4–8 mice per genotype, where each data point is a single mouse). Significance was determined by an unpaired two tailed t-test (mean ± s.e.m.). NS, not significant. c, Absolute numbers of B cell precursors (Hardy fraction A, B220+CD43+BP1−CD24−; Hardy fraction B, B220+CD43+BP1−CD24+; Hardy fraction C, B220+CD43+BP1+CD24+; Hardy fraction D, B220+CD43−IgM−IgD−; Hardy fraction E, B220+CD43−IgM+IgD−; Hardy fraction F, B220+CD43−IgM+IgD+) in the bone marrow (one femur and one tibia) (n = 4–8 mice per genotype, where each data point is a single mouse). Significance was determined by an unpaired two tailed t-test (mean ± s.e.m.). d, Splenic B cells cultured with the indicated stimuli (96 h) and stained for surface IgG1, IgE, IgG2b or IgG3 (n = 4–7 mice per genotype, where each data point is a single mouse). CSR 100%, mean immunoglobulin isotype switch frequency of two control animals in each experiment. Significance was determined by a two-way analysis of variance (ANOVA) with Tukey’s correction (mean ± s.e.m.). e, CTV-labeled splenic B cells were stimulated as indicated and stained for surface IgG1 after 96 h. Representative data, n > 6 mice. f, IgG1+ B cells as a proportion of total B cells (%) for each cell generation as determined by CTV staining and proliferation-associated dye dilution (n = 4–6 mice per genotype, where each data point is a single mouse; mean ± s.e.m.). g, CTV dilution in purified B cells cultured in the presence of LPS and IL-4 for 96 h. Representative data, n > 6 mice. h, NP-specific serum IgM (left) and IgG1 (right) at indicated times after NP-CGG immunization. Representative data, n = 2 independent experiments, each with four mice. Significance was determined by an unpaired two tailed t-test (mean ± s.e.m.). Gating strategies for the above flow cytometry data are provided in Supplementary Fig. 1.
Mutations affecting the CST gene CTC1 in humans cause Coats plus syndrome, an autosomal recessive disorder characterized by retinal telangiectasia, intracranial calcifications, osteopenia, gastrointestinal bleeding and, in severe cases, normocytic anaemia reflecting a degree of bone marrow failure33. Similarly, hematopoietic stem cell (HSC) exhaustion characterizes mice homozygous for Ctc1 gene deletions, leading to complete bone marrow failure and perinatal lethality34. International Mouse Phenotyping Consortium (IMPC)35 phenotyping of mice homozygous for a knockout-first conditional (ready) allele in Ctc1 (Ctc1tm1a(KOMP)Wtsi) also discovered perinatal lethality in Ctc1-deleted mice36. Thus, Ctc1 essentiality in murine hematopoiesis prompted us to investigate the effect of Ctc1 deletion in the B lymphocyte lineage, where the 53BP1 pathway supports normal development and programmed genome diversification. To this end, we generated conditional Ctc1tm1c/tm1c Mb1+/Cre mice (hereafter referred to as Ctc1F/F Mb1+/Cre mice), in which Cre expression from the Mb1-Cre transgene mediated complete deletion of Ctc1 critical exons 8–11 in early B cell progenitors (Fig. 1a and Extended Data Fig. 1c). To our surprise, CST deficiency was well tolerated in developing and differentiating B lymphocytes. Ctc1F/F Mb1+/Cre mice showed normal cell frequencies across all stages of lineage development in the bone marrow and B cell maturation in the spleen (Fig. 1b,c and Extended Data Fig. 1f). The normal development of Ctc1∆/∆ B cells aligned with the phenotypes of Shld2−/− and Shld3−/− mice, which similarly fostered normal B and T cell lineage development (Extended Data Fig. 1g–k). Normal lymphocyte development in shieldin-knockout mice is because of proficiency in V(D)J recombination, as demonstrated in our previous characterization of mice deleted of the shieldin gene Rev7 (ref. 11) and more recent concordant results in Shld1–Shld2-mutant mice12,13. Thus, neither CST nor shieldin contributes to lymphocyte development, where 53BP1 supports efficient DNA end joining during V(D)J recombination10,11.
Shieldin–CST cooperation underpins productive CSR
We next sought to define the importance of shieldin–CST interplay for the joining of AID-dependent DSBs during CSR, a process entirely dependent on 53BP1 that also requires the Rev7 and shieldin proteins11,12,13. To this end, we stimulated cultures of mature B splenocytes from Ctc1F/F Mb1+/Cre, Shld2−/− and Shld3−/− mice and analyzed their ability to support antibody isotype switching in vitro. Strikingly, ex vivo stimulation of mature splenic B cells from Ctc1F/F Mb1+/Cre mice revealed severe (>5-fold) defects in CSR across all analyzed IG isotypes, fully recapitulating the magnitude of defects presented in Shld2-deficient B cells analyzed in parallel (Fig. 1d,e). Defective class switching in Shld2-deficient and Ctc1-deficient B cells did not correlate to defects or differences in cell proliferation or survival (Fig. 1f,g). Despite this, B cells from Ctc1F/F Mb1+/Cre mice and both shieldin-knockout mouse strains supported higher class-switching frequencies than those from 53bp1−/− mice, where CSR was reduced >10-fold relative to WT (Fig. 1d,e and Extended Data Fig. 1l,m). These differences also held true in vivo in mice immunized with the model antigen NP-CGG (3-hydroxy-nitrophenyl acetyl coupled to chicken γ-globulin). Following immunization, serum titers of NP-specific IgG1 were strongly attenuated in Shld2−/−, 53bp1−/− and Ctc1F/F Mb1+/Cre mice relative to WT controls, despite exhibiting WT IgM-mediated antigen responses as expected (Fig. 1h and Extended Data Fig. 1n). However, antigen-specific IgG1 consistently accumulated to higher levels in Shld2−/− mice than in 53bp1−/− mice at all time points following immunization (Fig. 1h). By contrast, NP-specific IgG1 titers accumulated at equivalent levels following immunization in Ctc1F/F Mb1+/Cre and Shld2−/− mice (Extended Data Fig. 1n). Considered together, our results show that the 53BP1 pathway retains some capacity to support CSR in the absence of shieldin and CST. The fact that B lymphocyte development and differentiation capacities were found to be identical in Ctc1F/F Mb1+/Cre, Shld2−/− and Shld3−/− mice prompted us to next investigate the mechanistic basis of shieldin’s coordination with CST and downstream factors in CSR.
ATM promotes shieldin–CST interactions during DSB repair
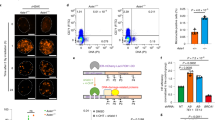
To investigate shieldin adaptor functions in DSB repair, we used proteomics to define the shieldin interactome. For this, we introduced TwinStrep-tagged Shld1 and Shld3 transgenes into Shld1−/− and Shld3−/− CH12-F3 mouse B cell lymphoma cell lines, respectively, and validated functional complementation at the level of rescued IgM-to-IgA class switching in stimulated cell cultures (Extended Data Fig. 2a,b). Because of shieldin’s extreme low abundance in mammalian cells26, its successful purification required lysates prepared from cultures of >6 × 109 cells. Shieldin complexes isolated by Strep-Tactin affinity purification were then subjected to analysis by liquid chromatography–tandem mass spectrometry (LC–MS/MS) over two independent experiments (Fig. 2a).
a, Schematic depicting shieldin purification and proteomic elucidation strategy. Briefly, Shld1/3-knockout CH12-F3 cell lines were lentivirally transduced with TwinStrep–HA–Shld1/3 espression transgenes. First, 300-ml suspension cultures were either mock-treated or irradiated (10 Gy) and lysates were prepared following a 1-h recovery. Shieldin complexes captured from lysates on MagStrep-XT resin were eluted in biotin, captured and tryptic-digested on S-Trap columns, with resulting peptides analyzed by LC–MS/MS. The schematic was generated using BioRender.com. b, B cell SHLD3 versus control (beads only) interactomes as defined by LC–MS/MS and LFQ. The scatter plot depicts the log2 fold enrichment of indicated MagStrep-XT purified complexes across two independent experiments. c, Comparison of mock and irradiated B cell SHLD3 interactomes as defined by LC–MS/MS and LFQ. The scatter plot depicts the log2 fold enrichment of indicated MagStrep-XT purified complexes across two independent experiments. d, Immunoblot analysis of CH12-F3 cell extracts from SHLD3 pulldown experiments. Representative of n = 2 independent experiments.
Highly-efficient purification of shieldin was confirmed by high peptide coverage across all four shieldin proteins (Table 1). Surprisingly, shieldin complexes were notably devoid of significant protein interactors (Fig. 2b). Because interactions between shieldin’s upstream regulators 53BP1 and RIF1 are controlled by DNA damage-dependent phosphorylation7,8, our subsequent investigation focused on SHLD1 and SHLD3 (SHLD1/3) complexes obtained from lysates of X-ray-irradiated cells.
Remarkably, analysis of (TwinStrep–SHLD1/3) purifications under these conditions revealed a discernible enrichment of peptides from CST proteins CTC1 and STN1 in both sets of purifications (Fig. 2c and Extended Data Fig. 2d). The induction of shieldin–CST interactions following DNA damage treatments was validated through immunoblotting of shieldin complex purifications on a smaller scale using anti-STN1 antibodies (Fig. 2d). Additionally, pretreatment of cells with a small-molecule ATM inhibitor (KU55933) but not an ataxia-telangiectasia and Rad3-related protein (ATR) inhibitor (AZD6738) abolished shieldin–CST interactions (Extended Data Fig. 2e).
These findings strongly implicate shieldin as a recruitment platform for the CST complex during DNA repair and point to ATM-dependent DNA damage signaling as the trigger for assembling shieldin–CST complexes at DSB sites. Such a conclusion provides a likely explanation for the equivalent effects of shieldin and CST gene disruption in murine B cells and, furthermore, implicates Pol α–primase-dependent fill-in synthesis as an intermediary step in the joining of AID-induced DSBs.
Shieldin-dependent DSB repair involves the reversionless 3-like (REV3L) Pol
During DNA replication, the synthesis of an RNA–DNA primer on the lagging strand by Pol α–primase precedes strand extension by DNA Pol δ37. We, therefore, considered that processive fill-in synthesis downstream of CST-directed Pol α–primase priming might likewise rely on Pol δ or an analogous processive DNA Pol. Interestingly, DNA Pol ζ, a translesion synthesis Pol formed of the B-family Pol REV3L in complex with REV7 and Pol δ accessory subunits POLD2 and POLD3 (ref. 38), was previously implicated in NHEJ during CSR39. This led us to investigate REV3L’s participation in DNA end joining downstream of shieldin–CST.
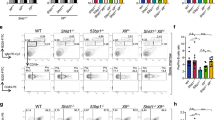
Rev3l is essential in mice38 and prior characterizations of B cell-specific REV3L functions were complicated by mosaicism driven by the out-competition of proliferation-defective Rev3l-deleted mature B cells with those that had escaped CD21-Cre-mediated recombination39. We, thus, opted to breed mice harboring the same Rev3ltm1(Rsky) conditional knockout allele in combination with the B cell-specific Mb1-Cre transgene, which mediates higher-efficiency Cre-mediated recombination upstream in B lymphocyte progenitor cells11,40. As predicted, Rev3l was completely deleted in the B cells of Rev3lF/F Mb1+/Cre mice (Extended Data Fig. 3a), where it resulted in 2–3-fold reductions in absolute B cell frequencies in the bone marrow and spleen (Fig. 3b). Despite this, purified B splenocytes from Rev3lF/F Mb1+/Cre mice were viable in culture and could undergo multiple rounds of cell division upon stimulation ex vivo (Fig. 3c). This allowed us to re-evaluate REV3L’s contribution to CSR. Indeed, Rev3lF/F Mb1+/Cre splenic B cells supported IgM-to-IgG1 class switching but at levels ~40% lower than WT or Mb1-cre-positive controls (Fig. 3d–f and Extended Data Fig. 3b). Again, class switching in cell-division-staged Rev3l-deficient B cells and their controls confirmed that these reductions were not a consequence of reduced cell proliferation (Fig. 3c,d), a result consistent with REV3L’s likely participation in NHEJ39.
a, Schematic representation of the Rev3l conditional allele. b, Absolute numbers of B220+ B cells in the bone marrow (one femur and one tibia) and spleen (n = 4–6 mice per genotype, where each data point is a single mouse). Significance was determined by an unpaired two tailed t-test (mean ± s.e.m.). c, CTV dilution in purified B cells cultured in the presence of LPS and IL-4 for 96 h. Representative data, n > 6 mice. d, IgG1+ B cells as a proportion of total B cells (%) for each cell generation as determined by CTV staining and proliferation-associated dye dilution (n = 4–6 mice per genotype, where each data point is a single mouse; mean ± s.e.m.). e, Splenic B cells cultured with the indicated stimuli (96 h) and stained for surface IgG1, IgE, IgG2b or IgG3 (n = 4–7 mice per genotype, where each data point is a single mouse). CSR 100%, mean immunoglobulin isotype switch frequency of two control animals in each experiment. Significance was determined by a two-way ANOVA with Tukey’s correction (mean ± s.e.m.). f, CTV-labeled splenic B cells were stimulated as indicated and stained for surface IgG1 after 96 h. Representative data, n > 6 mice. g, Splenic B cells cultured with the indicated stimuli (96 h) and stained for surface IgG1, IgE, IgG2b or IgG3 (n = 4–7 mice per genotype, where each data point is a single mouse). CSR 100%, mean immunoglobulin isotype switch frequency of two control animals in each experiment. Significance was determined by a two-way ANOVA with Tukey’s correction (mean ± s.e.m.). h, Splenic B cells cultured with the indicated stimuli (96 h) and stained for surface IgG1, IgE, IgG2b or IgG3 (n = 4–7 mice per genotype, where each data point is a single mouse). CSR 100%, mean immunoglobulin isotype switch frequency of two control animals in each experiment. Significance was determined by a two-way ANOVA with Tukey’s correction (mean ± s.e.m.).
To ascertain whether Pol ζ functions downstream of shieldin in CSR, we interbred Rev3lF/F Mb1+/Cre mice with Shld2-knockout mice and our previously described B cell conditional Rev7-knockout strain11, generating mice codeficient for shieldin and REV3L in the B cell lineage. The ex vivo stimulation of mature B splenocytes codeleted of Rev3l and either shieldin gene confirmed that Rev3l loss did not further decrease class switching below that seen in Rev7 or Shld2 single-knockout controls (Fig. 3g,h). This genetic epistasis between Rev3l and shieldin confirms Pol ζ’s participation in shieldin–CST-dependent DNA end-joining reactions. Considering CST’s role in catalyzing Pol α–primase-dependent fill-in synthesis in BRCA1-deficient cells and at Cas9-induced DSBs27,29,30, our results position Pol ζ as a likely mediator of processive fill-in synthesis downstream of these DNA priming reactions.
NHEJ-independent REV3L functions support B cell development
Despite the proliferative capacities of Rev3l∆/∆ Mb1+/Cre stimulated mature B splenocytes, the staging of B lineage cells in the bone marrow of Rev3lF/F Mb1+/Cre mice revealed developmental abnormalities. In these mice, diminished frequencies in pro B cell stage cells persisted throughout the small pre-B cell, immature B cell and mature circulating B cell stages and were noticeably more severe than those observed in 53bp1−/− mice (Fig. 4a and Extended Data Figs. 1h,n and 4a–c). This also contrasted with Rev7F/F Mb1+/Cre mice, where Mb1-cre-mediated deletion of Rev7 in B cells caused no development phenotypes (Fig. 4a)11, confirming that Pol ζ’s function in developing B cells requires its Pol REV3L but not its accessory subunit REV7. To determine the cause of B cell attrition in Revl3F/F Mb1+/Cre mice, we generated Rev3lF/F p53F/F Mb1+/Cre mice (Extended Data Fig. 4d). p53 deletion is known to suppress DNA damage-dependent apoptosis in murine lymphocytes41; likewise, its disruption rescued B cell frequencies in the bone marrow and spleen of Rev3lF/F p53F/F Mb1+/Cre mice (Fig. 4b,c and Extended Data Fig. 4e–g). By contrast, the expression of a transgenic B cell receptor specific for hen egg lysosome (MD4+/Tg) did not rescue B cell frequencies in Rev3lF/F Mb1+/Cre mice but did so in a V(D)J recombination-defective 53bp1−/− background (Fig. 4d and Extended Data Fig. 4h). On this basis, we surmise that, in the Rev3l-deleted background B cell lineage, abnormalities are most likely precipitated by replication-associated DNA damage and not by defects in V(D)J recombination. Thus, Pol ζ supports B lymphocyte development in a REV7-independent and DNA end-joining-independent manner. The rescue of B cell development conferred by p53 deletion is consistent with Pol ζ’s known role in suppressing DNA replication-associated genomic stability42, providing new evidence of REV7-independent functions of Pol ζ in genome maintenance.
a, Absolute numbers of B cell precursors (Hardy fraction A, B220+CD43+BP1−CD24−; Hardy fraction B, B220+CD43+BP1−CD24+; Hardy fraction C, B220+CD43+BP1+CD24+; Hardy fraction D, B220+CD43−IgM−IgD−; Hardy fraction E, B220+CD43−IgM+IgD−; Hardy fraction F, B220+CD43−IgM+IgD+) in the bone marrow (one femur and one tibia) (n = 4–6 mice per genotype, where each data point is a single mouse). Significance was determined by an unpaired two tailed t-test (mean ± s.e.m.). b, Absolute numbers of B220+ B cells in the bone marrow (one femur and one tibia) and spleen (n = 6–12 mice per genotype, where each data point is a single mouse). Significance was determined by an unpaired two tailed t-test (mean ± s.e.m.). c, Absolute numbers of B cell precursors in the bone marrow (one femur and one tibia) (n = 6–12 mice per genotype, where each data point is a single mouse). Significance was determined by an unpaired two tailed t-test (mean ± s.e.m.). d, Absolute numbers of mature IgM+IgD+ B cells in the spleen (n = 4 mice per genotype, where each data point is a single mouse). Significance was determined by an unpaired two tailed t-test (mean ± s.e.m.).
Limited role for shieldin in Brca1-deficient mouse embryos
The above findings confirm that the 53BP1 pathway directs multiple mechanistically distinct activities, which can be distinguished by shieldin–CST involvement. Synergy between these activities is required for the efficient joining of AID-induced DSBs during CSR; however, during V(D)J recombination, 53BP1 acts independently of shieldin, CST or REV3L. We reasoned that these activities might, therefore, each contribute separately to the pathological events in Brca1-deficient mice and tumors, where the 53BP1 pathway impedes HR and, through shieldin and CST, mediates toxic NHEJ events11,16,21,22,24,26,27,29. To test this, we interbred 53bp1 and Shld2 double-knockout mice with mice harboring Brca1 exon 5–13 deletions (Brca1+/−) that ablate all BRCA1 protein expression43. Brca1−/− mouse embryos die early in development, with none outlasting embryonic day 7.5 (E7.5)44. However, embryonic lethality can be circumvented upon deletion of 53bp1, yielding viable, albeit tumor-prone mice19,20. In our hands, Brca1−/−53bp1−/− mice were born at around half of the expected frequency (Fig. 5a and Table 2) and weighed less than their Brca1+/−53bp1−/− littermates, with most succumbing to thymic lymphoma between 12 and 20 weeks of age, consistent with recent reports20,45. By contrast, no Brca1−/−Shld2−/− double-knockout pups were born to intercrosses between Brca1+/−Shld2−/− mice (Table 2). Furthermore, we recovered only non-viable Brca1−/−Shld2−/− embryos between stages E8.5-E11.5, all of which exhibited severe developmental delay (Fig. 5b and Table 2). Of note, from staged embryos obtained in Brca1 heterozygous intercrosses performed in parallel (analyzed at E8.5 and E10.5), no Brca1−/− embryos were recovered. This implies that, unlike the case for 53bp1 deficiency, shieldin inactivation cannot restore viability in Brca1-null mice and only modestly delays the onset of embryonic lethality.
a, Representative image of 53bp1−/−Brca1−/− mice and relevant controls at ~12 weeks. Genotypes are indicated. b, Representative image of a Shld2−/−Brca1+/− control embryo and Shld2−/−Brca1−/− double-knockout embryo at E10.5. c, Survival of the indicated MEF cell lines grown for 7 days in the presence of the indicated doses of olaparib (n = 3 biological experiments; mean ± s.d.). d, Immunofluorescence microscopy of Rad51 IRIF in indicated MEF cell lines. Cells were irradiated (5 Gy) and fixed 2 h later. Representative images of n = 3 biological experiments. e, Quantification of RAD51 IRIF in Edu+ cells from d. Integrated intensity and foci quantifications were made using CellProfiler. Boxes indicate the 25th–75th percentiles with the median denoted and whiskers indicate the 10th–90th percentiles. Significance was determined by a two-sided Kruskal–Wallis H test with Dunn’s correction for multiple comparisons. ****P < 0.0001 and **P < 0.005 (n = 3 biological experiments). f, Survival of the indicated BARD1AID/AID HCT-116 cell lines grown for 7 days in the presence or absence of IAA (1 mM), doxycycline (2 mg ml−1) and the indicated doses of olaparib (n = 3 biological experiments; mean ± s.d.). g, Immunofluorescence microscopy of Rad51 IRIF in indicated BARD1AID/AID cell lines. Cultures were supplemented with doxycycline (2 mg ml−1 for 24 h) before the addition of IAA (1 mM). After 24 h, cells were irradiated (5 Gy) and fixed 2 h later. Representative images of n = 3 biological experiments. h, Quantification of RAD51 IRIF in Edu+ cells from g. Integrated intensity and foci quantifications were made using CellProfiler. Boxes indicate the 25th–75th percentiles with the median denoted and whiskers calculated by the Tukey method. Significance was determined by a two-sided Kruskal–Wallis H test with Dunn’s correction for multiple comparisons (n = 4 biological experiments).
DSB repair pathway choice control is shieldin independent
As shieldin loss blocked CSR but could not rescue the development of Brca1-null embryos, we hypothesized that the pro-NHEJ and HR-suppressive activities of the 53BP1 pathway might be distinguished at the level of shieldin involvement. Suggestive of 53BP1 operating independently of shieldin during HR suppression, we were unable to derive viable embryonic fibroblast (MEF) or embryonic stem cell (mESC) cell lines from Brca1−/−Shld2−/− E8.5 embryos and blastocyst outgrowths, respectively. We, therefore, resorted to addressing shieldin’s involvement in engineered BARD1AID/AID HCT-116 cells where BRCA1 can be conditionally inactivated through auxin degron-mediated degradation of its obligate binding partner protein BARD1 (refs. 14,15). Direct binding of DSB-proximal nucleosomes ubiquitinated on H2A Lys15 (H2AK15ub) by BARD1 is a prerequisite for BRCA1-dependent pathway choice control because it directly inhibits 53BP1–chromatin interactions at DNA damage sites14,17. Accordingly, in auxin-treated BARD1AID/AID HCT-116 cells (hereforth referred to as BARD1Δ/Δ cells), HR is fully restored upon deletion of 53BP1, resulting in a dramatic (albeit incomplete) suppression of cellular hypersensitivity to PARPi olaparib14. Mirroring the case in BARD1∆/∆ HCT-116, mouse embryonic fibroblast cell lines derived from Brca1−/−53bp1−/− embryos exhibited similar levels of residual olaparib sensitivity (Fig. 5c) yet WT levels of RAD51 recruitment upon irradiation (as visualized with two different RAD51 antibodies; Fig. 5d,e and Extended Data Fig. 5a,b), again indicative of HR restoration. These consistent results in murine and human cell lines reassured us of the suitability of BARD1AID/AID HCT-116 as a cellular model to investigate and compare the contributions of 53BP1 and shieldin to BRCA1 deficiency-associated HR defects.
In line with previous results in BRCA1-deficient cells11,22,26, deletion of SHLD2 or SHLD3 in BARD1Δ/Δ cells markedly enhanced cell survival in olaparib cytotoxicity assays. Importantly, however, olaparib resistance was less penetrant in BARD1Δ/ΔSHLD3−/− and BARD1Δ/ΔSHLD2−/− cells, as evidenced by the enhanced relative survival of BARD1Δ/Δ53BP1−/− cells in these experiments (Fig. 5f and Extended Data Fig. 5c–e). HR defects in replicating (S phase staged) BARD1Δ/ΔSHLD3−/− cells and BARD1Δ/ΔSHLD2−/− cells were also evident at the level of defective RAD51 recruitment into ionizing radiation-induced foci (IRIF), in stark contrast to BARD1Δ/Δ53BP1−/− cells, where RAD51 IRIF accumulated at WT (non-auxin-treated BARD1AID/AID cells) levels (Fig. 5g,h and Extended Data Fig. 5f,g). Furthermore, consistent differences in RAD51 recruitment were observed in irradiated KB1P-G3-derived Brca1−/−trp53−/− murine mammary tumor cell lines polyclonally deleted for either 53bp1 (ref. 46) or shieldin genes Shld2 (ref. 22) or Shld3 (ref. 11) (Extended Data Fig. 5h,i).
The different extents to which inactivation of 53BP1 or shieldin could suppress DNA repair defects in BRCA1-deficient BARD1Δ/Δ cells was also evident at the level of chromosome stability. In metaphase chromosome spreads from BARD1Δ/Δ cells, olaparib treatment induced high frequencies of chromosome or chromatid breaks and radial chromosomes (Fig. 6a,b and Extended Data Fig. 6). Chromosome breaks and radials were only slightly reduced in frequency in BARD1Δ/ΔSHLD3−/− cells, highlighting 53BP1’s retained capacity to block repair and mediate toxic joining events independently of shieldin. By contrast, both classes of chromosome lesion were nearly completely suppressed in BARD1Δ/Δ53BP1−/− cells (Fig. 6a,b), consistent with their restored capacity for HR (Fig. 5f,g and Extended Data Fig. 5f,g). We can, therefore, conclude that HR inactivation in BRCA1–BARD1-deficient cells is more profoundly dictated by 53BP1 status than the status of shieldin or its cooperating proteins. Nevertheless, our results show that, in the absence of BRCA1-dependent HR promotion, shieldin-directed DNA end joining exacerbates DNA damage-induced chromosomal instability, where it also makes substantial contributions to the overall hypersensitivity of cancer cells to HR deficiency-targeting PARPis (Fig. 6c).
a, Representative metaphase images from the indicated BARD1AID/AID cell lines. Cultures were supplemented with doxycycline (2 mg ml−1for 24 h) before the addition of IAA (1 mM). After a further 24 h, cells were treated with 500 nM olaparib and fixed 24 h later. Representative images of n = 3 biological experiments. b, Quantification of 40–65 metaphases per genotype per experiment. Significance was determined by a two-sided Kruskal–Wallis H test with Dunn’s correction for multiple comparisons. Horizontal bars indicate the mean values ± 95% confidence interval. ****P < 0.0001, ***P < 0.0005, **P < 0.005 and *P < 0.05 (n = 3 biological experiments). c, Model for 53BP1-dependent DSB repair. Chromatin-associated 53BP1 complexes suppress HR and promote V(D)J recombination. DNA damage-dependent signaling by RIF1 is conferred to shieldin through interactions with SHLD3. DSB signaling by ATM stimulates the assembly of shieldin–CST complexes on ssDNA, promoting the recruitment of Pol α–primase to initiate fill-in synthesis. Pol ζ REV3L increases the processivity of Pol α–primase-catalyzed fill-in synthesis.
Discussion
In this study, we adopted a genetic approach to systematically dissect the 53BP1 pathway. By comparing the traits of mice deficient in 53BP1, shieldin or CST, we aimed to elucidate the intricate relationship between 53BP1 and its downstream DNA-processing components in both normal and pathological contexts. Our findings shed light on the remarkable capability of 53BP1 to facilitate DSB repair even in the absence of shieldin. However, they also establish the indispensability of shieldin–CST interplay for efficient 53BP1-dependent DNA end joining during CSR and, furthermore, confirm Pol ζ’s involvement in these important DNA synthesis-dependent DNA end-joining reactions.
Shieldin is a DSB repair-specific mediator of CST
Upon binding ssDNA at telomeres and during DNA repair, CST acts as a catalyst of Pol α–primase-dependent DNA synthesis29,47. The discovery of shieldin–CST interdependence during CSR, thus, underscores the specialized nature of fill-in DNA synthesis as a prerequisite for the joining of AID-dependent DSBs. We additionally demonstrate that 53BP1’s reliance on shieldin–CST differs dramatically across different repair contexts. We posit that these differences reflect the unique DNA end-processing demands inherent to the spectrum of DSB structures generated in each distinct scenario, coupled with their varying frequencies of occurrence.
These conclusions are supported by multiple parallel observations in lymphocytes, mice and cancer cell models. In addition to the equivalent severity of class-switching defects in mature B cells from Shld2−/−, Shld3−/− and Ctc1F/F Mb1+/cre mice, hypothesis-agnostic elucidation of the shieldin interactome in B cells identified CST proteins as its primary interaction partners. Interestingly, interactions between shieldin and CST are induced upon DNA damage, a behavior explained by its reliance on the DSB-responsive kinase ATM. Upstream of shieldin, ATM-dependent phosphorylation of consensus Ser-Glu-containing motifs in 53BP1 promote the recruitment of RIF1 (refs. 7,48) by a recently discovered phospho-ligand-binding module in the RIF1 heat repeats8. Given the absence of additional shieldin interactors in our proteomics datasets, the ATM-dependent phosphorylation events that guide shieldin-dependent CST recruitment to DNA damage sites most likely involve a cryptic phospho-ligand-binding module within either shieldin or CST. From a structural perspective, it is noteworthy that neither CSR nor the ATM-dependent shieldin–CST interaction discovered here relies on a recently described N-terminal CTC1-binding motif in SHLD1 (ref. 29). Indeed, we found that a SHLD1∆exon1 protein deleted of its first 59 amino acids (encompassing this motif at aa 18–21) efficiently retrieved CST from lysates when they were prepared from irradiated B cells (Extended Data Fig. 2f). Similarly, mature splenic B cells from Shld1∆exon1 mice (a gift from the MMRRC49) confirmed to express an analogous truncated protein were also found to support near-WT class-switching frequencies in ex vivo stimulation experiments (Extended Data Fig. 2g,h), consistent with our previous results in Shld1−/− CH12-F3 cells expressing a Shld1∆N transgene29. Nevertheless, ATM-dependent shieldin–CST interactions are similarly consistent with shieldin’s importance in recruiting CST to DSB termini, explaining their interdependence in downstream DNA end joining.
Interestingly, this concept draws intriguing parallels with the regulation of CST-dependent C-strand fill-in synthesis at telomeres, governed by the shelterin complex50. In the telomeric context, the recruitment of CST to telomeres hinges on direct interactions between CST and the shelterin protein protection of telomere 1b (POT1b)47. Recent structural findings revealed that this involves a multisite interaction between CTC1 and regions within the triple OB-fold domain in POT1, which is further potentiated by POT1 phosphorylation51. A similar triple OB-fold architecture is also present in SHLD2 (refs. 11,21,22,26); thus, it is tempting to speculate that a similar mode of binding between CTC1 and structurally related features in POT1 or SHLD2 might underpin the initiation of fill-in DNA synthesis at telomeres and DSBs, respectively27,29,47.
Pol ζ acts downstream of shieldin–CST during DNA end joining
Using mice codeleted for Rev3l and either Shld2 or Rev7, we additionally detected an epistatic relationship between these genes in class-switching mature B cells. This offers a rational mechanistic explanation for Pol ζ’s role in NHEJ39, positioning it downstream of shieldin–CST, most likely in the extension of DNA repair synthesis that follows priming by Pol α–primase. Class switching in Rev3l-deficient B cells occurs at ~50% of WT frequencies, a level ~3–4-fold higher than that seen in those lacking shieldin genes or Ctc1. Does this indicate that Pol ζ is not the sole DNA Pol acting downstream of CST–Pol α–primase? As the typical extender of lagging-strand primers during DNA replication, Pol δ would be a likely redundant Pol. It is also possible that RNA–DNA primer synthesis on a short 5′ resected DNA end might be sufficient to stimulate the ligation of ssDNA-tailed DSBs by NHEJ, a pathway capable of ligating DSBs with ribonucleotide-incorporated termini52. Nevertheless, AID’s action during CSR generates ssDNA-tailed DSBs within switch (S) regions in the Igh locus, G+C-rich repeat-dense intronic elements abundant in palindromic motifs. The propensity of S-region ssDNA to form stem loops and/or parallel four-stranded G-quartets is well documented and such properties are speculated to contribute to various aspects of S-region function during CSR53. The formation of secondary DNA structures in the ssDNA-tailed termini of AID-dependent DSBs might similarly impede DNA synthesis by the canonical lagging-strand Pol δ, warranting the utility of a versatile TLS Pol to mediate DNA fill-in synthesis at Igh DSBs. Pol ζ also limits DNA damage accumulation during normal replication42, a function that we suggest extends to developing B cells that surprisingly does not involve REV7. Whether Pol ζ participates in other CST–Pol α–primase-catalyzed fill-in reactions remains to be explored. Such an interplay might be advantageous during C-strand fill-in at telomeres, where secondary DNA structures on the template G-strand could pose similar challenges to Pol δ processivity54.
53BP1 promotes DNA end joining independently of shieldin–CST
On the basis of the genetic analyses described above, we propose a refined model for the shieldin-dependent branch of the 53BP1 pathway (Fig. 6c). While vital for optimal CSR, the residual switching capacity of stimulated shieldin–CST-deficient B splenocytes notably exceeds that in 53bp1-deficient B cells, confirming that 53BP1 promotes the repair of a small subset of AID breaks without the requirement for fill-in synthesis. An additional paradigm for shieldin-independent NHEJ is V(D)J recombination, where 53BP1 supports the development of normal frequencies of B and T lymphocytes by enhancing long-range joining events10,11,13. Why does 53BP1 operate independently of shieldin–CST during V(D)J recombination? The simple explanation for this is RAG-induced DNA breaks have no utility for shieldin–CST-dependent fill-in synthesis yet profit from a distinct activity exerted by 53BP1. Our comparative investigations of 53BP1 and shieldin functions in BRCA1-deficient cells provide additional evidence in support of this proposition. Shieldin inactivation confers a degree of PARPi resistance to BARD1-depleted cells, yet resistance was further increased by deletion of 53BP1. Similar distinctions were perhaps hinted at by murine tumor studies. The potent survival benefit conferred by PARPi treatments in mice implanted with Brca1−/−Trp53−/− mammary tumors was completely lost when tumours were additionally deleted of 53bp1 (ref. 55); however, they modestly extended lifespan in equivalent tumor engraftment experiments when the tumors were instead deleted for Shld1 or Shld2 (ref. 22). Our knockout experiments in BARD1∆/∆ cells also revealed 53BP1 as the predominant mediator of PARPi-induced chromosome breaks and radial chromosomes, with shieldin making a lesser, albeit substantial and important contribution, in agreement with previous findings11,21,22,26,27,28,29. Given the different contributions that 53BP1 and shieldin make to the repair of RAG-induced and AID-induced DSBs in lymphocytes, we speculate that DSBs resulting from PARPi-induced replication blocks might likewise comprise a spectrum of DNA end structures, of which only a subset necessitate shieldin–CST axis-dependent repair.
HR suppression is a shieldin-independent function of 53BP1
One of the most striking deviations in phenotypes conferred by 53BP1 and shieldin losses was at the level of HR modulation in BRCA1–BARD1-deficient human and mouse cell lines. Here, 53BP1 deletion fully restored RAD51 recruitment to WT levels, indicative of fully competent HR. By contrast, the profound RAD51 recruitment defects that characterized human BARD1∆/∆ HCT-116 cells and murine KB1P tumor cells were only slightly ameliorated in shieldin-inactivated derivative cell lines. This distinction implies that, while DSB repair pathway choice regulation is dictated by 53BP1, it is only modestly influenced by downstream shieldin–CST-mediated DNA end processing. The weak degree of observed phenotypic suppression in Shld2−/−Brca1−/− embryos offers the most compelling support for this conclusion. Shieldin disruption only slightly delayed the onset of embryonic lethality, in stark contrast to the viability and superficially normal postnatal development of 53bp1−/−Brca1−/− mice. These findings may seem in contradiction to a recent report, where Shld2 knockout rescued embryonic and postnatal development in mice homozygous for the hypomorphic Brca1∆11 mutation12. However, these differences are almost certainly attributable to the notable residual functionality of the BRCA1∆11 protein. BRCA1∆11–BARD1 complexes are readily recruited to sites of DNA damage and retains BRCA1 functional domains including its coiled coil that links it to PALB2 (partner and localizer of BRCA2)–BRCA2 (ref. 56). However, DNA end resection is severely reduced in cells deleted of exon 11 in BRCA1 (ref. 56), a defect that might interfere with HR when challenged with antagonistic 53BP1-directed shieldin–CST fill-in activities.
In summary, our study reveals a surprising segregation of duties between 53BP1 and its downstream effector proteins during DSB repair. Shieldin–CST-dependent fill-in synthesis at DNA ends constitutes an ancillary reaction that supports the joining of DSBs with recessed ssDNA termini, such as those generated following the processing of staggered DNA cleavages induced by AID during CSR. Outside of CSR, however, 53BP1-dependent DSB repair remains remarkably resilient to loss of these effectors, consistent with a direct stimulation of DNA repair by chromatin-bound 53BP1 complexes. Our results also implicate upstream 53BP1 complexes, but not their DNA-processing effectors, as the major determinants of HR suppression in mammalian cells (Fig. 6c). This concept aligns well with our recently proposed chromatin-centric model for DSB pathway choice control, where HR and NHEJ outcomes are dictated by a mutual antagonism between BRCA1–BARD1 and 53BP1, upon their engagement of related DSB-proximal chromatin signatures14,15. Future work will be needed to address whether this hierarchy in the 53BP1 pathway will also be reflected in the clinical penetrance of mutations affecting 53BP1 and its effector genes in therapy-resistant BRCA1-mutant cancers.
Methods
Mice
The production and breeding of the genetically altered mice was carried out in accordance with the UK Home Office Animal (Scientific Procedures) Act 1986, with procedures reviewed by the Clinical Medicine Animal Welfare and Ethical Review Body at the University of Oxford, and conducted under project license PP8064604. Animals were housed in individually ventilated cages, provided with food and water ad libitum and maintained on a 12-h light–dark cycle (150–200 lx). The only reported positives on Federation of European Laboratory Animal Science Associations health screening over the entire time course of these studies were for Helicobacter hepaticus and Entamoeba spp. Experimental groups were determined by genotype and were, therefore, not randomized, with no animals excluded from the analysis. Sample sizes for fertility studies were selected on the basis of previously published studies and all phenotypic characterization was performed blind to the experimental group. All mice used in this study were generated on or backcrossed onto a C57BL/6 background (>5 generations). F1 mice harboring the Shld2tm1a (Shld2tm1a(EUCOMM)Hmgu, MGI: 5428631) allele were generated by backcrossing chimeric F0 mice generated by microinjection of gene-targeted C57BL/6N (JM8F6) derived mESC cells into albino C57BL6/J blastocysts. The targeting was performed in house using the targeting vector from the European Conditional Mouse Mutagenesis Program for the International Mouse Knockout Consortium, following the consortium’s recommended screening conditions. The resulting Shld2tm1a mice were then crossed with Flp-Cre-positive mice (Tg(ACTB-Flpe)9205Dym; Jax stock 005703) to generate a conditional allele with LoxP sites flanking exons 4 and 5, which includes the translation start site35. A targeted Shld2-null allele was subsequently generated by germline recombination of the LoxP sites using PGK-cre57 (B6.C-Tg(Pgk1-cre)1Lni/CrsJ; Jax stock 020811) (Fig. 1a). Shld3-null mice were generated at MRC Harwell by microinjection of Cas9 ribonucleoprotein complexes targeting exon 2 in the C57BL/6 background. Two founder mice, Shld3DEL2061−EM1 and Shld3DEL2061-EM2, with deletions of the sole coding exon, were generated (2,054 bp and 2,061 bp, respectively) (Fig. 1a). Helmholtz Munich/German Research Center for Environmental Health provided the Ctc1tm1a(KOMP)Wtsi mutant mouse line (allele: C57BL/6N-Ctc1<tm1a(KOMP)Wtsi>/Ieg). INFRAFRONTIER/EMMA (www.infrafrontier.eu (ref. 58)) and Helmholtz Zentrum Munich/German Research Center for Environmental Health distributed the sperm that was used to rederive the mouse line (EM: 10068) in our host institution’s transgenics core facility (MRC WIMM). Conditional Ctc1F/F mice were generated by crossing the ‘knockout-first’ allele (Ctc1tm1a(KOMP)Wtsi/Ieg; MGI: 4363331) to Flp-Cre-positive mice (Tg(ACTB-Flpe)9205Dym; Jax stock 005703) to generate mice with the Ctc1tm1c conditional allele. Experimental Ctc1F/F Mb1+/Cre, p53F/F Mb1+/Cre (Trp53floxed; MGI: 98834) and Rev3LF/F Mb1+/Cre (Rev3ltm1Rsky; MGI: 1337131) mice were generated by intercrossing with mice expressing the B cell lineage Mb1-cre deleter strain (Cd79atm1(cre)Reth; MGI:368745)40. The 53bp1−/− mice (Trp53bp1tm1Jc; MGI: 2654201) and Brca1−/− mice (Brca1null; MGI:104537) were generated as described elsewhere19,43,59. Shld1∆exon1 mice (C57BL/6NCrl-1110034G24Rikem1(IMPC)Mbp/Mmucd; stock 043862-UCD), obtained from the Mouse Biology Program (University of California, Davis) harbored a clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9-generated deletion of the terminal 71 bp of Shld1 exon 1. Complementary DNA sequencing confirmed the expression of a functional spliced Shld1∆exon1 transcript, initiated at an indel-generated in-frame ATG codon within the preserved frame-shifted sequences in exon 1 (ATG encoded within Tyr29–Glu30 codons).
All B cell phenotyping experiments involved age-matched 8–16-week-old male or female animals on an inbred C57BL/6 background. We did not pursue lower-penetrance phenotypes; thus, statistically significant data could typically be obtained with small group sizes (typically 4–10 mice). Randomization of samples was only undertaken during the scoring of chromosomal aberrations during metaphase analyses. Mice of a certain genotype were selected on the basis of a unique mouse identifier that does not indicate mouse genotype; thus, phenotype–genotype relationships were determined only at the data analysis stage. All experiments were approved by the University of Oxford Ethical Review Committee and performed under a UK Home Office Licence in compliance with animal use guidelines and ethical regulations.
Immunizations
WT, 53bp1−/−, Shld2−/−, Shld3−/− and Ctc1F/F Mb1+/cre mice were immunized intraperitoneally with 50 mg of NP-CGG (Santa Cruz Biotechnologies) emulsified in Imject Alum adjuvant (Pierce, Thermo Fisher). Blood samples were collected from the tail vein at 0, 7, 14, 21 and 28 days after immunization.
ELISA
ELISAs were used to quantify the production of NP-specific antibodies in mice serum. First, 96-well plates were coated with 1 μg ml−1 NP-BSA (Biosearch Technologies) in bicarbonate buffer, blocked with 5% milk in PBS and incubated with serial dilutions of serum collected at different time points from immunized mice. Plates were probed using alkaline phosphatase-coupled antibodies against mouse IgM and IgG1 (Southern Biotech). Phosphatase substrate (Sigma) was used for detection and optical density was measured at 405 nm. For IgG1, pooled blood from postimmunization WT mice was used as a standard and serially diluted into a standard curve. The first dilution was established as 1,000 arbitrary units (AU). For IgM, pooled blood from day 7 was used as a standard. Immunoglobulin concentrations in mouse serum or culture supernatants were determined by sandwich ELISA. Total IgG, IgM and IgA was measured with mouse IgG, IgM and IgA ELISA kits, respectively (Bethyl Laboratories), according to the manufacturer’s instructions. Mouse serum with known immunoglobulin concentrations of each immunoglobulin was used as a standard.
Micronuclei analysis
Micronuclei from peripheral blood samples were analyzed as described by Balmus et al.32. Approximately 50 μl of tail-vein blood was collected into heparin solution and immediately fixed in cold methanol. Samples were stored at −80 °C until processing. Cells were then stained with CD71 (1:400; 113805, BioLegend) and 1 μl (1 mg ml−1) propidium iodide.
Lymphocyte analysis and flow cytometry
Cell suspensions from bone marrow (one femur and one tibia), spleen and thymus were counted on a Pentra ES60 Hematology Analyzer (Horiba) and stained with anti-mouse antibodies against the following antigens as appropriate (all from BioLegend unless otherwise stated) in fluorescence-activated cell sorting (FACS) buffer (PBS with 2% BSA and 0.025% sodium azide): IgD (1:500; 405716 or Thermo Fisher 12-5993-82; 11-26c.2a), IgG1 (1:200; 406606, RMG1-1), IgM (1:500; 406506 or 406514, RMM-1), B220 (1:500; 103232, 103244, 103206 or 103212, RA3-6B2), BP1 (1:200; 108308, 6C3), CD4 (1:500; 100430), CD8a (1:500; 100732), CD19 (1:500; 115534 or 115520, 6D5), CD24 (1:1,000; 101827 or BD Pharmingen 562563, M1/69), CD93 (1:200; 136510, AA4.1), CD23 (1:200; BD Pharmingen 553139, B3B4), CD25 (1:500, 102008), CD43 (1:200; BD Pharmingen 562865, S7 or Thermo Fisher 11-0431-85, eBioR2/60), CD44 (1:200; 103022) and CD21 (1:500; BD Biosciences 563176, 7G6). Mouse BD Fc block (1:500, BD Pharmingen 553141) was added to block nonspecific binding and live–dead cells were discriminated after staining with Zombie Aqua viability dye (1:200; 423102). Data were acquired on an FACSCanto (BD Biosciences), LSR Fortessa X20 (BD Biosciences) or Attune NxT flow cytometer (Thermo Fisher) and were analyzed with FlowJo software version 10 (Tree Star). Gating strategies for bone marrow Hardy fractions are shown in Supplementary Fig. 1 and Extended Data Fig. 4.
Ex vivo B splenocyte culture, stimulation and flow cytometry
B cells were purified from red blood cell-lysed single-cell suspensions of mouse spleens by magnetic negative selection using a B cell isolation kit (Miltenyi Biotec, 130-090-862). B cells (3 × 105 per well in a 12-well plate) were cultured in RPMI supplemented with 10% fetal calf serum (FCS), 100 U per ml penicillin, 100 ng ml−1 streptomycin, 2 mM l-glutamine, 1× MEM nonessential amino acids, 1 mM sodium pyruvate and 50 μM β-mercaptoethanol. B cells were stimulated with 5 μg ml−1 lipopolysaccharide (LPS; Sigma, L7770-1MG), 10 ng ml−1 mouse recombinant interleukin (IL)-4 (Peprotech, 214-14-20) and agonist anti-CD40 antibody (0.5 μg ml−1; Miltenyl Biotec, FGK45.5). Cultures were grown at 37 °C with 5% CO2 under ambient oxygen conditions. Then, 4 days after seeding, stimulated B cells were analyzed using an FACSCanto or LSR Fortessa X20; analysis was performed using FlowJo. Cells were resuspended in FACS buffer, blocked with Mouse BD Fc block and immunostained with biotinylated antibodies as follows: anti-mouse IgG1 (1:100; BD Pharmingen 553441, A85-1), anti-mouse IgG2b (1:100; BioLegend 406704, RMG2b-1), anti-mouse IgG3 (1:100; BD Pharmingen 553401, R40-82) and streptavidin APC (1:500; Thermo Fisher 17-4317-82). Cells expressing IgE were assessed using anti-mouse IgE PE (1:200; BioLegend, 406908, RME-1). Live–dead cells were discriminated after staining with Zombie Aqua viability dye. Cell proliferation was assessed using CellTrace violet (CTV) according to the manufacturer’s instructions (CellTrace, Life Technologies).
Proteomics and MS
Pellets collected from cultures of ~4 × 108 CH12-F3 cells ± 10-Gy irradiation and 1-h recovery, were lysed in BLB (benzonase lysis buffer: 20 mM HEPES pH 7.9, 40 mM KCl, 2 mM MgCl2, 100 mM MEM, 10% glycerol, 0.5% NP-40, 50 U per ml benzonase (Novagen), 0.05% (v/v) phosphatase inhibitors (Sigma-Aldrich) and protease inhibitors (Complete EDTA-free, Roche)) and were incubated on ice for 30 min before a second incubation with adjusted salt (450 mM KCl). Flag–REV7 or control complexes were isolated from clarified lysates, following their dilution in NSB (no-salt buffer: 20 mM HEPES pH 7.9, 10% glycerol, 0.5 mM DTT, 0.5 mM EDTA, 0.05% (v/v) phosphatase inhibitors (Sigma-Aldrich) and protease inhibitors (Roche)) to a final salt concentration of 125 mM. TwinStrep–HA–SHLD1/3 complexes were immunopurified on MagStrep-XT magnetic beads (IBA), washed extensively in wash buffer (BLB supplemented with 100 mM KCl and 0.1% NP-40) and eluted with 50 μM biotin and snap-frozen for downstream proteomics. For western blot analysis, MagStep XT beads were then mixed with Laemmli buffer and boiled at 95 °C for 5 min before loading on SDS–PAGE gels. Protein samples were fractionated on NuPAGE 4–12% 1.0 mM Bis-Tris polyacrylamide gels (Life Technologies, NP0322) before transferring to 0.45-μm nitrocellulose membranes (GE Healthcare, 10600003). After transfer, membranes with blocked with 5% milk in PBST for at least 30 min and then incubated overnight with primary antibody in PBST supplemented with 0.03% NaN3 and 3% BSA. Primary antibodies used for western blot in this study included rabbit anti-53BP1 (1:2,500; Novus Biological, NB100-304), rabbit anti-SHLD1/2/3 (1:250; in-house), mouse anti-REV7 (1:500; BD, 612266), mouse anti-STN1 (1:500; Santa Cruz, sc-376450), mouse anti-tubulin (1:10,000; Sigma, 00020911) and rabbit anti-phospho-Chk1 (Ser345, 1:500; Cell Signaling, 2348). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (Vector Laboratories, PI-1000) and HRP-conjugated horse anti-mouse IgG (Vector Laboratories, PI-2000) secondary antibodies were used and membranes developed with Clarity Western enhanced chemiluminescence substrate (Bio-Rad, 170-5061) or SuperSignal West Pico PLUS (Thermo Fisher) and imaged using a Gel Doc XR System (Bio-Rad). Images were then opened and exported for publication using Image Lab.
TwinStrep peptide eluted complexes were solubilized in 4% (w/v) SDS and 50 mM TEAB, reduced and alkylated using DTT and iodoacetamide and digested with trypsin on S-Trap microcolumns as per the manufacturer’s instructions. Samples were analyzed on an LC–MS platform (Ultimate 3000 nHPLC and Q-Exactive HFX MS instrument). Peptides were separated on an 50-cm EASY-Spray column with a gradient of 3–35% acetonitrile in 5% DMSO and 0.1% formic acid at 250 nl min−1. MS1 spectra were acquired at a resolution of 60,000 at 200 m/z. Up to the 15 most abundant precursor ions were selected for subsequent MS/MS analysis after ion isolation with a mass window of 1.6 Th. Peptides were fragmented by HCD with 27% collision energy. Raw files were analyzed by Progenesis QI (version 3, Waters) for spectral label-free quantitation (LFQ) with default parameters (top-three quantitation mode). Proteins were identified with PEAKS 8.0 (Bioinformatics Solutions) using standard parameters and the UniProt mouse reviewed proteome (retrieved January 23, 2020). The peptide and protein false discovery rate was set to 1%.
Cell lines, cell culture and CRISPR–Cas9 editing
All CH12-F3 cell lines were cultured in RPMI supplemented with 5% NCTC-109 medium, 10% FCS, 100 U per ml penicillin, 100 ng ml−1 streptomycin and 2 mM l-glutamine at 37 °C with 5% CO2 under ambient oxygen conditions. Rev7−/−, c20orf196−/− (Shld1−/−) and Flj26957−/− (Shld3−/−) CH12-F3 cells were generated using CRISPR–Cas9 as previously described11. Complemented cell lines were generated by lentivirus-mediated transduction, using viral supernatants collected from 293T cells cotransfected with third-generation packaging vectors and a pLenti-PGK-TwinStrep-Flag-HA-PURO-DEST vector containing cloned transgene inserts. Typically, cells were spinoculated with viral supernatants supplemented with polybrene (8 μg ml−1) and HEPES (20 mM) (1,500 r.p.m., 90 min at 25 °C). Stable cell lines were subsequently selected and maintained in the presence of puromycin (1 μg ml−1). To stimulate CSR to IgA, CH12-F3 cells were stimulated with agonist anti-CD40 antibody (0.5 μg ml−1; Miltenyi Biotec; FGK45.5), mouse IL-4 (5 ng μl−1; R&D Systems) and transforming growth factor-β1 (2.5 ng μl−1; R&D Systems). Cell-surface IgA expression was determined by flow cytometric staining with anti-mouse IgA–PE antibody (Cambridge Bioscience; 1040-09).
E12.5 MEFs prepared and SV40 LargeT-immortalized by standard procedures were used in all experiments unless otherwise indicated. All MEF cell lines were maintained in high-glucose DMEM (Sigma-Aldrich, D6546) supplemented with 10% FBS, penicillin–streptomycin and 2 mM l-glutamine.
BARD1AID/AID and BARD1AID/AID53BP1−/− cell lines were generated as previously described14,15. All BARD1AID/AID and derivative cell lines were maintained in high-glucose DMEM (Sigma-Aldrich, D6546) supplemented with 10% FBS, penicillin–streptomycin and 2 mM l-glutamine. Cultures were maintained at 37 °C with 5% CO2. BARD1AID/AIDSHLD2−/− and BARD1AID/AIDSHLD3−/− knockout cell lines were generated by CRISPR–Cas9 as previously described14. Gene-specific guide RNAs were integrated into pSpCas9(BB)-2A-GFP (PX458) (Addgene, 48138) and 2 μg of plasmid was electroporated into 106 cells using a Lonza 4D-Nucleofector according to the manufacturer’s protocol for HCT-116 cells. Green fluorescent protein-positive cells were sorted 24 h after electroporation using a Sony SH800 cell sorter with the brightest 5% being pooled for recovery in medium containing 50% FBS for 4 days. Sorted populations were then seeded at low density and individual clones were isolated after 10 days of outgrowth. Individual clones were validated by sequencing.
The KB1P-G3 (ref. 46) Brca1−/−p53−/− mouse mammary tumor cell lines were cultured in DMEM in the presence of 10% FCS and penicillin–streptomycin (Gibco) under low-oxygen conditions (3% O2, 5% CO2 at 37 °C). KB1P-G3 Shld2−/− Cl.1 and 2 and KB1P-G3B1 (Brca1-reconstituted) cells were kindly gifted from S. Rottenberg22. KB1P-G3 Shld3−/− cells were generated by CRISPR–Cas9 as previously described11.
Survival experiments
To generate survival curves for and MEF cell lines, seeding assays were first performed on 104 cells allowed to grow for 7 days. Following this, the growth medium was removed and the cells were washed briefly with PBS before the addition of crystal violet stain (0.5% crystal violet in 25% methanol). Cells were stained for 5 min, washed with double-distilled H2O and dried before scanning. Cell seeding numbers were calculated relative to WT cell density. Survival curves were then generated by seeding 300–400 WT or Brca1+/−53bp1−/− cells or 650–750 Brca1−/−53bp1−/− cells in triplicate for each drug concentration in a 96-well plate. Olaparib was added 24 h later to the indicated final concentrations.
Survival curves for BARD1AID/AID and derivative cell lines were generated as previously described14. Briefly, 300/1000 (DMSO/IAA condition) BARD1AID/AID cells, 300/700 BARD1AID/AID53BP1−/− cells or 400/1500 BARD1AID/AIDSHLD2/3−/− cells, per well were seeded in the presence of doxycycline (2 μg ml−1) in triplicate for each drug concentration in a 96-well plate. After 24 h, IAA (1 mM) or carrier (DMSO) was added. Then, 1 h after IAA addition, olaparib was added to the indicated final concentrations.
For both cell lines, 7 days after drug addition, the medium was replaced with phenol red-free DMEM (Thermo Fisher, 21063-029) supplemented with 10% FBS, penicillin–streptomycin, 2 mM l-glutamine and 10 μg ml−1 resazurin (Sigma-Aldrich, R7017). Plates were then returned to the incubator for 2–4 h or until the growth medium in untreated control wells began to develop a pink color. Relative fluorescence was measured with a BMG LABTECH CLARIOstar plate reader. The mean of three technical repeats after background subtraction was taken as the value for a biological repeat and three biological repeats were performed for each experiment. All survival curves presented in this article represent the mean of three biological repeats ± s.e.m.
Immunofluorescence
For RAD51 foci quantification in BARD1AID/AID and derivative cell lines, 2 × 105 cells were passed through a 70-μm mesh cell strainer and seeded on three fibronectin-coated glass coverslips (13 mm) in a single well of a six-well plate in the presence of doxycycline (2 μg ml−1). After 24 h, IAA was added to a final concentration of 1 mM. Cells were irradiated (5 Gy) 2 h after IAA addition and fixed in 2% PFA 2 h after irradiation. For RAD51 foci quantification in MEFs and KB1P-G3 cells, 4 × 105 and 5 × 105 cells, respectively, were seeded on three glass coverslips (13 mm) in a single well of a six-well plate. Cells were irradiated (5 Gy) and fixed in 2% PFA 2 h after irradiation. Staining of all fixed cells began with 15-min blocking (3% BSA and 0.1% Triton X-100 in PBS) followed by 1-h incubation with primary antibody in a humidity chamber. For experiments in which cells were treated with EdU, the Click-iT EdU cell proliferation kit (Alexa Fluor 647; Thermo Fisher, C10340) was used to label EdU-positive cells according to the manufacturer’s protocol between blocking and primary antibody incubation.
The following primary antibodies were used at the indicated concentrations: rabbit anti-RAD51 (HCT-116: 1:1,000; 70-001 BioAcademia, MEFS and KB1P-G3: 1:2,000; ab133534 Abcam or 1:1,000; anti-RAD51 rabbit serum, a gift from R. Kanaar) and mouse anti-γH2AX (1:500; 05-636 Millipore). Following primary antibody introduction, coverslips were washed three times with PBS containing 0.1% Triton X-100 before incubation with secondary antibody for 1 h in a humidity chamber. Secondary antibodies used in this study were as follows: goat anti-mouse Alexa Fluor 488 (1:500; A-11001 Invitrogen) and goat anti-rabbit Alexa Fluor 568 (1:500; A-11011 Invitrogen). Coverslips were then washed three times with PBS containing 0.1% Triton X-100, washed once with PBS and mounted on glass microscope slides using a drop of ProLong Gold antifade reagent with DAPI (Life Technologies, P36935).
Images were acquired on a Leica DMi8 wide-field microscope. CellProfiler (Broad Institute) was used for foci quantification. Images were visualized and saved in Fiji and assembled into figures in Affinity Designer.
Cytological analysis
Metaphase spreads were prepared by standard methods. In brief, detached cells were resuspended in 75 mM KCl for 20 min before being fixed in Carnoy’s fixative. Approximately 20 μl of cell suspension was dropped onto clean slides and left to dry overnight. The cells were then stained with 0.5 μg ml−1 propidium iodide in PBS for 20 min and rinsed before the slides were mounted in Vectashield and DAPI (Vector Labs). The slides were analyzed blind and a minimum of 40 metaphases were acquired using an Olympus BX60 microscope for epifluorescence equipped with a Sensys charge-coupled device camera (Photometrics). Images were collected using Genus Cytovision software (Leica).
Statistics
Prism 10 (GraphPad software) was used for graphing and statistical analysis. Relevant statistical methods for individual experiments are detailed within figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The MS proteomics data were deposited to the ProteomeXchange Consortium through the PRIDE partner repository with the dataset identifier PXD045534. Raw immunofluorescence images and .fcs files are available upon request. Source data are provided with this paper.
References
Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Wang, X. S., Lee, B. J. & Zha, S. The recent advances in non-homologous end-joining through the lens of lymphocyte development. DNA Repair (Amst.) 94, 102874 (2020).
Scully, R., Panday, A., Elango, R. & Willis, N. A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 20, 698–714 (2019).
Saha, T., Sundaravinayagam, D. & Di Virgilio, M. Charting a DNA repair roadmap for immunoglobulin class switch recombination. Trends Biochem. Sci. 46, 184–199 (2021).
Hu, Q. et al. Mechanisms of BRCA1–BARD1 nucleosome recognition and ubiquitylation. Nature 596, 438–443 (2021).
Fradet-Turcotte, A. et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54 (2013).
Chapman, J. R. et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell 49, 858–871 (2013).
Setiaputra, D. et al. RIF1 acts in DNA repair through phosphopeptide recognition of 53BP1. Mol. Cell 82, 1359–1371 e9 (2022).
Setiaputra, D. & Durocher, D. Shieldin—the protector of DNA ends. EMBO Rep. 20, e47560 (2019).
Difilippantonio, S. et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 456, 529–533 (2008).
Ghezraoui, H. et al. 53BP1 cooperation with the REV7–shieldin complex underpins DNA structure-specific NHEJ. Nature 560, 122–127 (2018).
Ling, A. K. et al. SHLD2 promotes class switch recombination by preventing inactivating deletions within the Igh locus. EMBO Rep. 21, e49823 (2020).
Vincendeau, E. et al. SHLD1 is dispensable for 53BP1-dependent V(D)J recombination but critical for productive class switch recombination. Nat. Commun. 13, 3707 (2022).
Becker, J. R. et al. BARD1 reads H2A lysine 15 ubiquitination to direct homologous recombination. Nature 596, 433–437 (2021).
Nakamura, K. et al. H4K20me0 recognition by BRCA1–BARD1 directs homologous recombination to sister chromatids. Nat. Cell Biol. 21, 311–318 (2019).
Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010).
Chapman, J. R., Sossick, A. J., Boulton, S. J. & Jackson, S. P. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J. Cell Sci. 125, 3529–3534 (2012).
Lord, C. J. & Ashworth, A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat. Med. 19, 1381–1388 (2013).
Chen, J. et al. 53BP1 loss rescues embryonic lethality but not genomic instability of BRCA1 total knockout mice. Cell Death Differ. 27, 2552–2567 (2020).
Nacson, J. et al. BRCA1 mutation-specific responses to 53BP1 loss-induced homologous recombination and PARP inhibitor resistance. Cell Rep. 24, 3513–3527 (2018).
Dev, H. et al. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat. Cell Biol. 20, 954–965 (2018).
Noordermeer, S. M. et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature 560, 117–121 (2018).
Xu, G. et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521, 541–544 (2015).
Barazas, M. et al. The CST complex mediates end protection at double-strand breaks and promotes PARP inhibitor sensitivity in BRCA1-deficient cells. Cell Rep. 23, 2107–2118 (2018).
Findlay, S. et al. SHLD2/FAM35A co-operates with REV7 to coordinate DNA double-strand break repair pathway choice. EMBO J. 37, e100158 (2018).
Gupta, R. et al. DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell 173, 972–988 (2018).
Mirman, Z. et al. 53BP1–RIF1–shieldin counteracts DSB resection through CST- and Pol α-dependent fill-in. Nature 560, 112–116 (2018).
Tomida, J. et al. FAM35A associates with REV7 and modulates DNA damage responses of normal and BRCA1-defective cells. EMBO J. 37, e99543 (2018).
Mirman, Z., Sasi, N. K., King, A., Chapman, J. R. & de Lange, T. 53BP1–shieldin-dependent DSB processing in BRCA1-deficient cells requires CST–Pol α–primase fill-in synthesis. Nat. Cell Biol. 24, 51–61 (2022).
Schimmel, J., Munoz-Subirana, N., Kool, H., van Schendel, R. & Tijsterman, M. Small tandem DNA duplications result from CST-guided Pol α–primase action at DNA break termini. Nat. Commun. 12, 4843 (2021).
Mirman, Z. & de Lange, T. 53BP1: a DSB escort. Genes Dev. 34, 7–23 (2020).
Balmus, G. et al. A high-throughput in vivo micronucleus assay for genome instability screening in mice. Nat. Protoc. 10, 205–215 (2015).
Anderson, B. H. et al. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat. Genet. 44, 338–342 (2012).
Gu, P. et al. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 31, 2309–2321 (2012).
Skarnes, W. C. et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 (2011).
Groza, T. et al. The International Mouse Phenotyping Consortium: comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res. 51, D1038–D1045 (2023).
Kunkel, T. A. & Burgers, P. M. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 18, 521–527 (2008).
Martin, S. K. & Wood, R. D. DNA polymerase ζ in DNA replication and repair. Nucleic Acids Res. 47, 8348–8361 (2019).
Schenten, D. et al. Pol ζ ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J. Exp. Med. 206, 477–490 (2009).
Hobeika, E. et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl Acad. Sci. USA 103, 13789–13794 (2006).
Lowe, S. W., Schmitt, E. M., Smith, S. W., Osborne, B. A. & Jacks, T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362, 847–849 (1993).
Lange, S. S., Wittschieben, J. P. & Wood, R. D. DNA polymerase ζ is required for proliferation of normal mammalian cells. Nucleic Acids Res. 40, 4473–4482 (2012).
Liu, X. et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc. Natl Acad. Sci. USA 104, 12111–12116 (2007).
Hakem, R. et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell 85, 1009–1023 (1996).
Li, M. et al. 53BP1 ablation rescues genomic instability in mice expressing ‘RING-less’ BRCA1. EMBO Rep. 17, 1532–1541 (2016).
Jaspers, J. E. et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov. 3, 68–81 (2013).
Cai, S. W. & de Lange, T. CST–Pol α/primase: the second telomere maintenance machine. Genes Dev. 37, 555–569 (2023).
Di Virgilio, M. et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339, 711–715 (2013).
Amos-Landgraf, J. et al. The Mutant Mouse Resource and Research Center (MMRRC): the NIH-supported national public repository and distribution archive of mutant mouse models in the USA. Mamm. Genome 33, 203–212 (2022).
Takai, H., Aria, V., Borges, P., Yeeles, J. T. P. & de Lange, T. CST–polymerase α–primase solves a second telomere end-replication problem. Nature 627, 664–670 (2024).
Cai, S. W., Takai, H., Walz, T. & de Lange, T. POT1 recruits and regulates CST-Pol α/primase at human telomeres. Cell 187, 3638–3651 (2024).
Pryor, J. M. et al. Ribonucleotide incorporation enables repair of chromosome breaks by nonhomologous end joining. Science 361, 1126–1129 (2018).
Chaudhuri, J. & Alt, F. W. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4, 541–552 (2004).
Lormand, J. D. et al. DNA polymerase δ stalls on telomeric lagging strand templates independently from G-quadruplex formation. Nucleic Acids Res. 41, 10323–10333 (2013).
Duarte, A. A. et al. BRCA-deficient mouse mammary tumor organoids to study cancer-drug resistance. Nat. Methods 15, 134–140 (2018).
Nacson, J. et al. BRCA1 mutational complementation induces synthetic viability. Mol. Cell 78, 951–959 (2020).
Lallemand, Y., Luria, V., Haffner-Krausz, R. & Lonai, P. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 7, 105–112 (1998).
INFRAFRONTIER ConsortiumINFRAFRONTIER—providing mutant mouse resources as research tools for the international scientific community. Nucleic Acids Res. 43, D1171–D1175 (2015).
Ward, I. M., Minn, K., van Deursen, J. & Chen, J. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23, 2556–2563 (2003).
Acknowledgements
We thank all members of the laboratory of J.R.C. for discussions, J. Jonkers (NKI) for Brca1−/− & trp53F/F mouse models, S. Rottenberg (University of Berne) and R. Kanaar (Erasmus MC) for sharing KB1P cell lines and RAD51 antibodies, S. Zha (Columbia University), B. Sleckman and B.-R. Chen (University of Alabama at Birmingham) for advice, reagents and cell lines, M. Deobagkar for technical assistance, the European Mouse Mutant Cell Repository (www.eummcr.info) and Mutant Mouse Resource and Research Centers (www.mmrrc.org) for mouse strains and gene-targeting constructs and the University of Oxford Department of Biomedical Services (BMS) Functional Genetics and JR Hospital BMS facilities for technical support. This work was funded by the Medical Research Council (MRC, UK) project grant MR/R017549/1 and MRC Molecular Hematology Unit grants MC_UU_00016/19 and MC_UU_00029/2. Salary to J.R.C. and funding for his group comes from a Cancer Research UK (CRUK) Senior Cancer Research Fellowship (RCCSCF-Nov21\100004) and previously Career Development Fellowship (C52690/A19270), which provided salary support to C.O. and J.R.B. A.K. and J.R.B. were previously supported by EMBO Long-Term (EMBO ALT 542-2020) and Ruth L. Kirschstein NRSA Individual Postdoctoral Fellowships (F32) (National Institutes of Health National Cancer Institute, F32CA239339), respectively. The IMPC supported this work through UK Research Institute grant MC_UP_1502/3. This work was previously supported by the Wellcome Trust (Core Award Grant, 203141/Z/16/Z). J.R.C. is the recipient of Lister Institute Research Prize Fellowship funding.
Author information
Authors and Affiliations
Contributions
Conceptualization, A.K. and J.R.C. Methodology, A.K., R.P., C.O., L.S, J.R.B., D.B., C.P., B.D. and J.R.C. Investigation, A.K., P.I.R., J.S.M., R.P., D.M., C.O. and B.D. Formal analysis, A.K. and R.P. Data curation, R.P. Writing—original draft, review and editing, A.K. and J.R.C. Funding acquisition, J.R.C. Supervision, A.K. and J.R.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Dimitris Typas, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Equivalent immunodeficiencies in mice lacking shieldin and CST.
a) Representative image of PCR amplicons from genomic DNA obtained by ear biopsies from mice of the indicated genotype. Bands of different size correspond to the Shld2tm1a allele (274 bp), Shld2tm1c allele (554 bp), Shld2tm1d allele (240 bp) and WT allele (391 bp). n = >20 mice per genotype. b) Representative image of PCR amplicons from genomic DNA obtained by ear biopsies from mice of the indicated genotype. Bands of different size correspond to the Shld3EM1-B6 allele (left) or Shld3EM2-B6 allele (right) (~312 bp) and WT allele (459 bp). n = >20 mice per genotype. c) Representative image of PCR amplicons from genomic DNA obtained by ear biopsies (left), splenic b cells or purified splenic b cells (right) from mice of the indicated genotype. Bands of different size correspond to the Ctc1fl allele (413 bp), Ctc1−/− allele (658 bp) and WT allele (210 bp). n = >20 mice per genotype except for purified B cells; n = 2 independent experiments each with 2-3 mice. d) Peripheral blood stained as indicated. Reticulocytes (RET), micronucleated reticulocytes (MN-RET), normochromatic erythrocytes (NCE) and micronucleated normochromatic erythrocytes (MN-NCE). Representative data, n ³ 5 mice of each sex per genotype. e) MN-NCE as a percentage of total NCEs from tail vein blood of mice at 8-12 weeks. n = 5-12 mice of each sex per genotype, where each data point is a single mouse. Significance was determined by unpaired two tailed t-test, Mean ± SEM. f) Flow cytometric sub-classification of nature splenic b cell fractions. n = 4-8 mice per genotype, where each data point is a single mouse. Significance was determined by unpaired two tailed t-test, Mean±SEM. g) Absolute numbers of B220+ B cells in the bone marrow (one femur and one tibia) and spleen and live lymphocytes in the thymus. n = 10-18 mice per genotype, where each data point is a single mouse. Significance was determined by unpaired two tailed t-test, Mean ± SEM. h) Absolute numbers of B cell precursors in the bone marrow (one femur and one tibia). n = 15-19 mice per genotype, where each data point is a single mouse. Significance was determined by unpaired two tailed t-test, Mean±SEM. i) Flow cytometric sub-classification of mature splenic b cell fractions. n = 6-17 mice per genotype, where each data point is a single mouse. Significance was determined by unpaired two tailed t-test, Mean ± SEM. j) Absolute numbers of thymic T-cells, n = 9-17 mice per genotype, where each data point is a single mouse. Mean ± SEM. k) Absolute numbers of T cell precursors in double negative (CD4- CD8-) in the thymus, n = 9-17 mice per genotype, where each data point is a single mouse. Mean ± SEM. l) CTV-labelled purified B cells were stimulated as indicated and stained for surface IgG/IgE on day 4. Representative of n > 6 experiments. m) Splenic B cells cultured with the indicated stimuli (96 h) and stained for surface IgG1, IgE, IgG2b or IgG3. n = 11-14 mice per genotype. CSR 100%, mean immunoglobulin isotype switch frequency of 2 control animals in each experiment. Significance was determined by two-way ANOVA with Tukey’s correction. Mean ± SEM. n) NP-specific serum IgM (left) and IgG1 (right) at indicated times after NP-CGG immunization. AU, arbitrary units. Representative data, n = 2 independent experiments, each with 2-3 mice. Significance was determined by unpaired two tailed t-test. Mean ± SEM.
Extended Data Fig. 2 DNA damage induces interactions between shieldin and CST.
a) IgM-to-IgA CSR frequencies in Shld3 knockout CH12-F3 cell lines, n = 3 independent experiments, where each data point is the average of 3 technical replicates from a single experiment. Mean ± SEM. Representative flow cytometry plots for IgM-to-IgA CSR. Representative data, n > 3 independent experiments. b) IgM-to-IgA CSR frequencies in Shld1 knockout CH12-F3 cell lines, n = 3 independent experiments, where each data point is the average of 3 technical replicates from a single experiment. Mean ± SEM. Representative flow cytometry plots for IgM-to-IgA CSR. Representative data, n > 3 independent experiments. c) B cell Shld1 interactome as defined by LC–MS/MS and LFQ. Scatter plot depicts log2 fold-enrichment of indicated TwinStep–immunocomplexes across 2 independent experiments. d) B cell Shld1 IR-dependent interactome as defined by LC–MS/MS and LFQ. Scatter plot depicts log2 fold-enrichment of indicated TwinStep–immunocomplexes across 2 independent experiments. e) Western blot analysis of CH12-F3 cell extracts from pulldown experiments. Cells were pre-treated with ATM, ATR inhibitor or both for 1 hr prior to irradiation. Representative of n = 2 independent experiments. f) Western blot analysis of CH12-F3 cell extracts from pulldown experiments. Representative of n = 2 independent experiments. g) Splenic B cells cultured with the indicated stimuli (96 h) and stained for surface IgG1, IgE, IgG2b or IgG3. n = 3-5 mice per genotype. CSR 100%, mean immunoglobulin isotype switch frequency of 2 control animals in each experiment. Significance was determined by two-way ANOVA with Tukey’s correction. Mean ± SEM. h) CTV-labelled purified B cells were stimulated as indicated and stained for surface IgG on day 4. Representative of n = 2 experiments.
Extended Data Fig. 3 Pol ζ cooperation with shieldin supports NHEJ in CSR.
a) Representative image of PCR amplicons from genomic DNA obtained by ear biopsies (left), splenic b cells or purified splenic b cells (right) from mice of the indicated genotype. Bands of different size correspond to the Rev3lfl allele (458 bp), Rev3l−/− allele (300 bp) and WT allele (384 bp). n = >20 mice per genotype except for purified B cells; n = 2 independent experiments each with 2-3 mice. b) CTV-labelled purified B cells were stimulated as indicated and stained for surface IgG/IgE on day 4. Representative of n > 5 experiments.
Extended Data Fig. 4 REV7-independent and NHEJ-independent functions of Pol ζ support B cell development.
a) Flow cytometric sub-classification of nature splenic b cell fractions. n = 4-6 mice per genotype, where each data point is a single mouse. Significance was determined by unpaired two tailed t-test, Mean±SEM. b) Flow cytometry analysis of B cell development in the bone marrow of Mb1+/Cre, Rev7f/lf Mb1+/Cre or Rev3lfl/fl Mb1+/Cre mice; gating on B220+CD43+ (top, Hardy fractions A, B and C) and on B220+CD43− (bottom, Hardy fractions D, E and F). Representative data; n = 3 experiments. c) Flow cytometry analysis of mature b cell populations in the spleen of Mb1+/Cre, Rev7f/lf Mb1+/Cre or Rev3lfl/fl Mb1+/Cre mice; gating on B220+CD19+. Representative data; n = 3 experiments. d) Representative image of PCR amplicons from genomic DNA obtained by ear biopsies (left), splenic b cells or purified splenic b cells (right) from mice of the indicated genotype. Bands of different size correspond to the p53fl allele (353 bp), p53−/− allele (450 bp) and WT allele (200 bp). n = >20 mice per genotype except for purified B cells; n = 2 independent experiments each with 2-3 mice. e) Flow cytometric sub-classification of nature splenic b cell fractions. n =6-12 mice per genotype, where each data point is a single mouse. Significance was determined by unpaired two tailed t-test, Mean±SEM. f) Flow cytometry analysis of B cell development in the bone marrow of Rev+/+ p53fl/fl Mb1+/Cre or Rev3lfl/fl p53fl/fl Mb1+/Cre mice; gating on B220+CD43+ (top, Hardy fractions A, B and C) and on B220+CD43− (bottom, Hardy fractions D, E and F). Representative data; n = 3 experiments. g) Flow cytometry analysis of mature b cell populations in the spleen of Rev+/+ p53fl/fl Mb1+/Cre or Rev3lfl/fl p53fl/fl Mb1+/Cre mice; gating on B220+CD19+. Representative data; n = 3 experiments. h) Flow cytometry analysis of B cell development in the bone marrow of MD4+/Tg, Rev3fl/fl Mb1+/Cre MD4+/Tg, 53BP1−/− MD4+/TG mice; gating on B220+. Representative data; n = 3 experiments.
Extended Data Fig. 5 53BP1-dependent HR suppression in BRCA1-deficient cells is shieldin independent.
a) Immunofluorescent microscopy of Rad51 IRIF in indicated MEF cell lines using Rabbit serum gifted by R. Kanaar. Cells were irradiated (5 Gy) and fixed 2 h later. Representative images of n = 3 biological experiments. b) Quantification of RAD51 IRIF in Edu+ cells from D. Integrated intensity and foci quantifications were made using CellProfiler. Boxes indicate the 25th–75th percentiles with the median denoted, and whiskers indicate the 10th-90th percentile. Significance was determined by two-sided Kruskal–Wallis H test with Dunn’s correction for multiple comparisons. p values; ****p < 0.0001, **p < 0.0005, **p < 0.005, *p < 0.05. n = 3 biological experiments. c) Survival of the indicated BARDAID/AID cell lines grown for 7 days in the presence of doxycycline (2 μg ml−1) and the indicated doses of olaparib, n = 3 biological experiments, Mean±SEM. d) Survival of the indicated BARD1AID/AID HCT-116 cell-lines grown for 7 days in the presence or absence of IAA (1mM), doxycycline (2 mg ml−1) and the indicated doses of Olaparib, n = 3 biological experiments, Mean±SD. e) Survival of the indicated BARD1AID/AID HCT-116 cell-lines grown for 7 days in the presence or absence of IAA (1mM), doxycycline (2 mg ml−1) and the indicated doses of Olaparib, n = 3 biological experiments, Mean±SD. f) Immunofluorescent microscopy of Rad51 IRIF in indicated BARD1AID/AID cell lines. Cultures were supplemented with doxycycline (2 mg ml−1 for 24 h) before addition of IAA (1 mM). After 24 h cells were irradiated (5 Gy) and fixed 2 h later. Representative images of n = 3 biological experiments. g) Quantification of RAD51 IRIF in Edu+ cells from F. Integrated intensity and foci quantifications were made using CellProfiler. Boxes indicate the 25th–75th percentiles with the median denoted, and whiskers calculated by the Turkey method. Significance was determined by two-sided Kruskal–Wallis H test with Dunn’s correction for multiple comparisons. p values; ****p < 0.0001, ***p < 0.0005, **p < 0.005, *p < 0.05. h) Immunofluorescent microscopy of Rad51 IRIF in indicated KB1P-G3 mouse mammary tumour cell lines. Cells were irradiated (5 Gy) and fixed 2 h later. Representative images of n = 3 biological experiments. i) Quantification of RAD51 IRIF in Edu+ cells from H. Integrated intensity and foci quantifications were made using CellProfiler. Boxes indicate the 25th–75th percentiles with the median denoted, and whiskers calculated by the Turkey method. Significance was determined by two-sided Kruskal–Wallis H test with Dunn’s correction for multiple comparisons. p values; ****p < 0.0001, ***p < 0.0005, **p < 0.005, *p < 0.05.
Extended Data Fig. 6 Independent and synergistic contributions of 53BP1 and shieldin to genomic instability in BRCA1-deficient cells.
a) Quantification of between 40 and 65 metaphases analysed per genotype/experiment. Cultures were supplemented with doxycycline (2 μg ml−1 for 24 h) before addition of IAA (1 mM). After a further 24 h cells were treated with DMSO and fixed 24 h later. Significance was determined by two-sided Kruskal–Wallis H test with Dunn’s correction for multiple comparisons. p values; ****p < 0.0001, ***p < 0.0005, **p < 0.005, *p < 0.05. All comparisons not indicated are not significant. n = 3 biological experiments.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Table 1A,B.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
King, A., Reichl, P.I., Metson, J.S. et al. Shieldin and CST co-orchestrate DNA polymerase-dependent tailed-end joining reactions independently of 53BP1-governed repair pathway choice. Nat Struct Mol Biol 32, 86–97 (2025). https://doi.org/10.1038/s41594-024-01381-9
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41594-024-01381-9
This article is cited by
-
Mechanisms and regulation of DNA end resection in the maintenance of genome stability
Nature Reviews Molecular Cell Biology (2025)
-
RIF1 integrates DNA repair and transcriptional requirements during the establishment of humoral immune responses
Nature Communications (2025)