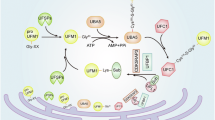
Abstract
The role of modification by ubiquitin-fold modifier (‘UFMylation’) in regulating metastasis has remained enigmatic. Cell migration, a critical step in metastasis, is driven by actin polymerization mediated by actin-related proteins 2 and 3 (Arp2/3) at the leading edge of lamellipodia. Here, we report that UFM1-specific E3 ligase 1 (UFL1) interacts with and catalyzes the UFMylation of ArpC4, a core subunit of the Arp2/3 complex. Akt has a key role in this process, which involves phosphorylating UFL1 at T426, thereby enhancing its interaction with ArpC4 and inducing ArpC4 UFMylation. Through ArpC4 UFMylation and potentially other targets, UFL1 facilitates lamellipodia formation and promotes cell migration, invasion and metastasis, making UFL1 an attractive therapeutic target for cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The protein structures used in this study were obtained from the PDB under accession codes 5IAA, 1K8K and 6W18. The data supporting the findings of this study are available within the article and Supplementary Information. Source data are provided with this paper.
References
Clark, A. G. & Vignjevic, D. M. Modes of cancer cell invasion and the role of the microenvironment. Curr. Opin. Cell Biol. 36, 13–22 (2015).
Krause, M. & Gautreau, A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 15, 577–590 (2014).
Goley, E. D. & Welch, M. D. The Arp2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7, 713–726 (2006).
Swaney, K. F. & Li, R. Function and regulation of the Arp2/3 complex during cell migration in diverse environments. Curr. Opin. Cell Biol. 42, 63–72 (2016).
Gournier, H., Goley, E. D., Niederstrasser, H., Trinh, T. & Welch, M. D. Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol. Cell 8, 1041–1052 (2001).
Robinson, R. C. et al. Crystal structure of Arp2/3 complex. Science 294, 1679–1684 (2001).
Goley, E. D. et al. An actin-filament-binding interface on the Arp2/3 complex is critical for nucleation and branch stability. Proc. Natl Acad. Sci. USA 107, 8159–8164 (2010).
Rotty, J. D., Wu, C. Y. & Bear, J. E. New insights into the regulation and cellular functions of the Arp2/3 complex. Nat. Rev. Mol. Cell Biol. 14, 7–12 (2013).
Dang, I. et al. Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature 503, 281–284 (2013).
Rocca, D. L., Martin, S., Jenkins, E. L. & Hanley, J. G. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat. Cell Biol. 10, 259–271 (2008).
Zuo, X. F. et al. Exo70 interacts with the Arp2/3 complex and regulates cell migration. Nat. Cell Biol. 8, 1383–1388 (2006).
Schnoor, M., Stradal, T. E. & Rottner, K. Cortactin: cell functions of a multifaceted actin-binding protein. Trends Cell Biol. 28, 79–98 (2018).
Cai, L., Makhov, A. M., Schafer, D. A. & Bear, J. E. Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell 134, 828–842 (2008).
Zhao, K. L. et al. WDR63 inhibits Arp2/3-dependent actin polymerization and mediates the function of p53 in suppressing metastasis. EMBO Rep. 21, e49269 (2020).
Zheng, S. H. et al. Role and mechanism of actin-related protein 2/3 complex signaling in cancer invasion and metastasis: a review. Medicine 102, e33158 (2023).
Molinie, N. & Gautreau, A. The Arp2/3 regulatory system and its deregulation in cancer. Physiol. Rev. 98, 215–238 (2018).
Keenan, K. E., Zachman, D. K. & Hirschey, M. D. Discovering the landscape of protein modifications. Mol. Cell 81, 1868–1878 (2021).
Dikic, I. & Schulman, B. A. An expanded lexicon for the ubiquitin code. Nat. Rev. Mol. Cell Biol. 24, 273–287 (2023).
Komatsu, M. et al. A novel protein-conjugating system for UFM1, a ubiquitin-fold modifier. EMBO J. 23, 1977–1986 (2004).
Kang, S. H. et al. Two novel ubiquitin-fold modifier 1 (UFM1)-specific proteases, UFSP1 and UFSP2. J. Biol. Chem. 282, 5256–5262 (2007).
Millrine, D. et al. Human UFSP1 is an active protease that regulates UFM1 maturation and UFMylation. Cell Rep. 40, 111168 (2022).
Tatsumi, K. et al. A novel type of E3 ligase for the UFM1 conjugation system. J. Biol. Chem. 285, 5417–5427 (2010).
Zhou, X. C. et al. UFMylation: a ubiquitin-like modification. Trends Biochem. Sci. 49, 52–67 (2024).
Peter, J. J. et al. A non-canonical scaffold-type E3 ligase complex mediates protein UFMylation. EMBO J. 41, e111015 (2022).
Tatsumi, K. et al. The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat. Commun. 2, 181 (2011).
Ishimura, R. et al. The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3. Nat. Commun. 13, 7857 (2022).
Qin, B. et al. UFL1 promotes histone H4 ufmylation and ATM activation. Nat. Commun. 10, 1242 (2019).
Wang, Z. F. et al. MRE11 UFMylation promotes ATM activation. Nucleic Acids Res. 47, 4124–4135 (2019).
Wang, L. H. et al. UFMylation of RPL26 links translocation-associated quality control to endoplasmic reticulum protein homeostasis. Cell Res. 30, 5–20 (2020).
Walczak, C. P. et al. Ribosomal protein RPL26 is the principal target of UFMylation. Proc. Natl Acad. Sci. USA 116, 1299–1308 (2019).
Liang, J. R. et al. A genome-wide ER-phagy screen highlights key roles of mitochondria! metabolism and ER-resident UFMylation. Cell 180, 1160–1177 (2020).
Zhang, M. et al. RCAD/Ufl1, a UFM1 E3 ligase, is essential for hematopoietic stem cell function and murine hematopoiesis. Cell Death Differ. 22, 1922–1934 (2015).
Cai, Y. F. et al. UFBP1, a key component of the UFM1 conjugation system, is essential for UFMylation-mediated regulation of erythroid development. PLoS Genet. 11, e1005643 (2015).
Yiu, S. P. T. et al. An Epstein–Barr virus protein interaction map reveals NLRP3 inflammasome evasion via MAVS UFMylation. Mol. Cell 83, 2367–2386 (2023).
Tao, Y. J. et al. UFL1 promotes antiviral immune response by maintaining STING stability independent of UFMylation. Cell Death Differ. 30, 16–26 (2023).
Snider, D. L., Park, M., Murphy, K. A., Beachboard, D. C. & Horner, S. M. Signaling from the RNA sensor RIG-I is regulated by ufmylation. Proc. Natl Acad. Sci. USA 119, e2119531119 (2022).
Lee, L. et al. UFMylation of MRE11 is essential for telomere length maintenance and hematopoietic stem cell survival. Sci. Adv. 7, eabc7371 (2021).
Gong, Y. et al. PARP1 UFMylation ensures the stability of stalled replication forks. Proc. Natl Acad. Sci. USA 121, e2322520121 (2024).
Wang, Z. et al. VCP/p97 UFMylation stabilizes BECN1 and facilitates the initiation of autophagy. Autophagy 20, 2041–2054 (2024).
Tian, T. et al. UFL1 triggers replication fork degradation by MRE11 in BRCA1/2-deficient cells. Nat. Chem. Biol. 20, 1650–1661 (2024).
Yoo, H. M. et al. Modification of ASC1 by UFM1 is crucial for ERα transactivation and breast cancer development. Mol. Cell 56, 261–274 (2014).
Liu, J. et al. UFMylation maintains tumour suppressor p53 stability by antagonizing its ubiquitination. Nat. Cell Biol. 22, 1056–1063 (2020).
Yang, J. J. et al. Metformin induces ferroptosis by inhibiting UFMylation of SLC7A11 in breast cancer. J. Exp. Clin. Cancer Res. 40, 206 (2021).
Zhou, J. Z. et al. Dysregulation of PD-L1 by UFMylation imparts tumor immune evasion and identified as a potential therapeutic target. Proc. Natl Acad. Sci. USA 120, e2306087120 (2023).
He, C. et al. UFL1 ablation in T cells suppresses PD-1 UFMylation to enhance anti-tumor immunity. Mol. Cell 84, 1120–1138 (2024).
Wang, K. et al. The ufmylation modification of ribosomal protein L10 in the development of pancreatic adenocarcinoma. Cell Death Dis. 14, 350 (2023).
Wanka, V., Fottner, M., Cigler, M. & Lang, K. Genetic code expansion approaches to decipher the ubiquitin code. Chem. Rev. 124, 11544–11584 (2024).
Shaaban, M., Chowdhury, S. & Nolen, B. J. Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state. Nat. Struct. Mol. Biol. 27, 1009–1016 (2020).
Luan, Q., Liu, S. L., Helgeson, L. A. & Nolen, B. J. Structure of the nucleation-promoting factor SPIN90 bound to the actin filament nucleator Arp2/3 complex. EMBO J. 37, e100005 (2018).
Kazazian, K. et al. Plk4 promotes cancer invasion and metastasis through Arp2/3 complex regulation of the actin cytoskeleton. Cancer Res. 77, 434–447 (2017).
LeClaire, L. L., Baumgartner, M., Iwasa, J. H., Mullins, R. D. & Barber, D. L. Phosphorylation of the Arp2/3 complex is necessary to nucleate actin filaments. J. Cell Biol. 182, 647–654 (2008).
LeClaire, L. L., Rana, M., Baumgartner, M. & Barber, D. L. The Nck-interacting kinase NIK increases Arp2/3 complex activity by phosphorylating the Arp2 subunit. J. Cell Biol. 208, 161–170 (2015).
Vadlamudi, R. K., Li, F., Barnes, C. J., Bagheri-Yarmand, R. & Kumar, R. p41-Arc subunit of human Arp2/3 complex is a p21-activated kinase-1-interacting substrate. EMBO Rep. 5, 154–160 (2004).
Montor, W. R., Salas, A. R. O. S. E. & de Melo, F. H. M. Receptor tyrosine kinases and downstream pathways as druggable targets for cancer treatment: the current arsenal of inhibitors. Mol. Cancer 17, 55 (2018).
Chen, M. M. et al. GPS 6.0: an updated server for prediction of kinase-specific phosphorylation sites in proteins. Nucleic Acids Res. 51, W243–W250 (2023).
Gerakis, Y., Quintero, M., Li, H. L. & Hetz, C. The UFMylation system in proteostasis and beyond. Trends Cell Biol. 29, 974–986 (2019).
Chung, C. H. & Yoo, H. M. Emerging role of protein modification by UFM1 in cancer. Biochem. Biophys. Res. Commun. 633, 61–63 (2022).
Jing, Y., Mao, Z. M. & Chen, F. L. UFMylation system: an emerging player in tumorigenesis. Cancers 14, 3501 (2022).
van der Kammen, R. et al. Knockout of the Arp2/3 complex in epidermis causes a psoriasis-like disease hallmarked by hyperactivation of transcription factor Nrf2. Development 144, 4588–4603 (2017).
Singh, S. et al. Identification of the p16-Arc subunit of the Arp 2/3 complex as a substrate of MAPK-activated protein kinase 2 by proteomic analysis. J. Biol. Chem. 278, 36410–36417 (2003).
Bogucka-Janczi, K. et al. ERK3/MAPK6 dictates CDC42/RAC1 activity and ARP2/3-dependent actin polymerization. eLife 12, e85167 (2023).
Papalazarou, V. & Machesky, L. M. The cell pushes back: the Arp2/3 complex is a key orchestrator of cellular responses to environmental forces. Curr. Opin. Cell Biol. 68, 37–44 (2021).
Enomoto, A. et al. Akt/PKB regulates actin organization and cell motility via girdin/APE. Dev. Cell 9, 389–402 (2005).
Manning, B. D. & Toker, A. Akt/PKB signaling: navigating the network. Cell 169, 381–405 (2017).
Revathidevi, S. & Munirajan, A. K. Akt in cancer: mediator and more. Semin Cancer Biol. 59, 80–91 (2019).
Glaviano, A. et al. PI3K/Akt/mTOR signaling transduction pathway and targeted therapies in cancer. Mol. Cancer 22, 138 (2023).
Islam, M., Jones, S. & Ellis, I. Role of Akt/protein kinase B in cancer metastasis. Biomedicines 11, 3001 (2023).
He, Y. et al. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target. Ther. 6, 425 (2021).
Gorelik, R. & Gautreau, A. Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc. 9, 1931–1943 (2014).
Pijuan, J. et al. In vitro cell migration, invasion, and adhesion assays: from cell imaging to data analysis. Front. Cell Dev. Biol. 7, 107 (2019).
Oweis, W. et al. Trans-binding mechanism of ubiquitin-like protein activation fevealed by a UBA5–UFM1 complex. Cell Rep. 16, 3113–3120 (2016).
Acknowledgements
We thank Y. Yin of the Mass Spectrometry System at the National Facility for Protein Science (Shanghai Advanced Research Institute, Chinese Academy of Science) for mass spectrometry sample preparation, data collection and data analysis. We also thank X. Yao for helpful discussion. This work was supported by grants from the National Natural Science Foundation of China (32270811 and 32470749, to Y.M.; 32200614, to K.Z.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB0940302, to Y.M.), the Center for Advanced Interdisciplinary Science and Biomedicine of IHM (QYPY20220006, to Y.M.) and the Youth Science and Technology Talents Support Program (2024) by Anhui Association for Science and Technology (RCTJ202408, to K.Z.). This work was also supported by the advanced computing resources provided by the Supercomputing Center of the USTC. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
K.Z. and Y.M. designed the research. K.Z., H.H., D.F., M.X., J.C., S.Z., S.T., M.W., X.G., N.Y., B.Y. and W.J. performed the research. K.Z., H.H., D.F., M.X., J.C., S.Z., S.T., M.W., X.G., N.Y., B.Y., W.J., C.W. and Y.M. analyzed the data. K.Z. and Y.M. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Alexis Gautreau, Krishnaraj Rajalingam and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editors: Jean Nakhle and Dimitris Typas, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 UFL1 promotes cell migration, invasion, and metastasis.
(a–c) A549 cells expressing control shRNA, UFL1 shRNA#1, or UFL1 shRNA#2 were subjected to wound-healing (b) and transwell migration (c) assays. The successful knockdown of UFL1 was verified by western blotting (a). Scale bar in b: 200 μm; c: 100 μm. (d, e) U2OS cells expressing control shRNA, UFL1 shRNA#1, or UFL1 shRNA#2 were subjected to transwell migration assays (e). The successful knockdown of UFL1 was verified by western blotting (d). Scale bar in e: 100 μm. (f, g) A549 cells expressing control shRNA, UFL1 shRNA#1, or UFL1 shRNA#2 were subjected to single cell tracking (f) and transwell invasion (g) assays. Cell migration speed, persistence, and mean-squared displacement (MSD) in single cell tracking assay were also analyzed using an Excel macro described by Gorelik and Gautreau (f). Scale bar in f: 25 μm; g: 100 μm. (h) U2OS cells expressing control shRNA, UFL1 shRNA#1, or UFL1 shRNA#2 were subjected to transwell invasion assays. Scale bar: 100 μm. (i–k) H1299 expressing control sgRNA, UFL1 sgRNA#1, or UFL1 sgRNA#2 were subjected to transwell migration (j) and transwell invasion (k) assays. Cell lysates were also analyzed by western blotting (i). Scale bar in j and k: 100 μm. All experiments were repeated three times independently with similar results. The shown blots and images are representative of three independent experiments. Data in (b, c, e, g, h, j, k) are mean ± SD (n = 3). Data in (f) are mean ± SEM (n = 10, 8 and 10, respectively). Statistical analysis was performed using one-way ANOVA (b, c, e–h, j, k).
Extended Data Fig. 2 UFL1 promotes lamellipodia formation.
(a–d) A549 cells with or without ectopic Flag-UFL1 expression were serum-starved for 6 h before they were stimulated with or without 10% FBS or 100 ng/ml EGF for 30 min, followed by immunostaining with either anti-UFL1 (a) or anti-Flag (c) antibody, along with FITC-phalloidin. The plot profiles (a, c) and Pearson’s correlation coefficient (b, d) were quantified. The arrows indicate the leading edge of migrating cells. Scale bar: 20 μm. (e–h) H1299 cells (e, f) or A549 cells (g, h) expressing control shRNA, UFL1 shRNA#1, or UFL1 shRNA#2 were serum-starved for 6 h before they were scratched. Cells were then treated with 100 ng/ml EGF (e, f) or 10% FBS (g, h) for 30 min, followed by immunofluorescence assay. (e, g) The lamellipodia protrusions driven by branched actin networks were shown by immunostaining with anti-Cortactin antibody. White arrows indicate lamellipodia region. (f, h) The percentage of lamellipodia-positive cells were calculated. Scale bar: 20 μm. (i) A549 cells expressing control shRNA, UFL1 shRNA#1, or UFL1 shRNA#2 were subjected to cell spreading assay. Scale bars: 50 μm. (j) H1299 cells expressing control shRNA, UFL1 shRNA#1, or UFL1 shRNA#1 plus shRNA-resistant Flag-UFL1 were subjected to cell spreading assay. Scale bars: 50 μm. All experiments were repeated three times independently with similar results. The shown images are representative of three independent experiments. Data in (b, d, f, h, i, j) are mean ± SD (n = 3). Statistical analysis was performed using one-way ANOVA (f, h–j).
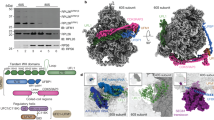
Extended Data Fig. 3 ArpC4 is a bona fide substrate of UFMylation.
(a) Lysates from HEK293T cells expressing Flag-UFL1 alone or together with the indicated HA-tagged each single subunit of the Arp2/3 complex were immunoprecipitated with anti-Flag antibody, followed by western blot analysis. (b) Lysates from HEK293T cells expressing HA-UFL1 alone or HA-UFL1 plus Flag-tagged Arp3, Cortactin, Coronin, and Cofilin as indicated were immunoprecipitated by anti-Flag antibody, followed by western blot analysis. (c) Lysates from HEK293T cells expressing HA-UFL1 alone or HA-UFL1 plus Flag-tagged N-WASP were immunoprecipitated by anti-Flag antibody, followed by western blot analysis. (d) Myc-tagged UBA5 and UFC1, HA-tagged UFL1, UFBP1, and UFM1-ΔC2 were expressed in HEK293T cells with Flag-tagged ArpC4 as indicated. Denatured cell lysates were subjected to pull-down with anti-Flag antibody, followed by western blot analysis. (e) ArpC4, UFC1, UFL1, and UFBP1, together with UFM1-ΔC2 or UFM1-ΔC3, were expressed in HEK293T cells. Denatured cell lysates were subjected to pull-down with Ni-NTA resins, followed by western blot analysis. (f) Flag-tagged ArpC4 and the UFMylation components were expressed in HEK293T cells with or without HA-tagged UFSP2. Denatured cell lysates were subjected to pull-down with anti-Flag antibody, followed by western blot analysis. (g) His-tagged wild-type ArpC4 or its KO mutant (all lysine residues were replaced with arginine) was expressed in HEK293T cells with the UFMylation components as indicated. Denatured cell lysates were subjected to pull-down with Ni-NTA resins, followed by western blot analysis. (h) UFSP2-KO H1299 cells expression control or Flag-UFM1-ΔC2 were infected with lentiviruses expressing either control shRNA or ArpC4 shRNA as indicated. Denatured cell lysates were subjected to pull-down with anti-Flag antibody, followed by western blot analysis. (i) Coomassie blue staining of recombinant His-tagged UBA5, UFC1, UFM1-ΔC2, UFL1/UFBP1, and MBP-tagged ArpC4. All experiments were repeated three times independently with similar results. The shown blots/gels are representative of three independent experiments.
Extended Data Fig. 4 Mass spectrometric identification of lysine residues of ArpC4 modified by UFM1.
(a–g) Recombinant MBP-ArpC4 and the UFMylation components were subjected to in vitro UFMylation assay. The products were separated by SDS-PAGE, followed by commassie blue staining. The band of UFMylated ArpC4 was cut and delivered to mass spectrometry analysis. Lys35 (a), Lys44 (b), Lys60 (c), Lys77 (d), Lys84 (e), Lys107 (f), and Lys166 (g) of ArpC4 were identified as the potential UFMylation sites.
Extended Data Fig. 5 ArpC4 UFMylation plays an important role in cell migration, invasion, and metastasis.
(a–c) Control or ArpC4 knockdown A549 cells were infected with lentiviruses expressing Flag-tagged ArpC4 or its 5KR mutant. Cells were subjected to wound-healing (a) and transwell migration (c) assays. Cell lysates were also analyzed by western blot (b). Scale bar in a: 200 μm; c: 100 μm. (d–f) The analysis of cell migration speed (d), persistence (e), and mean-squared displacement (MSD) (f) of Fig. 4d. (g) Control or ArpC4 knockdown A549 cells were infected with lentiviruses expressing Flag-tagged ArpC4 or its 5KR mutant. Cells were subjected to transwell invasion assays. Scale bar: 100 μm. (h) H1299 cells expressing either control shRNA or ArpC4 shRNA were infected with lentiviruses expressing Flag-tagged wild-type ArpC4 or its 5KR mutant. Cells were serum-starved for 6 h before they were scratched. Cells were then treated with 10% FBS for 30 min, followed by immunofluorescence assay. The percentage of lamellipodia-positive cells were calculated. (i) H&E staining of lung lobes from the NCG mice intravenously injected with A549 cells expressing control shRNA, ArpC4 shRNA, ArpC4 shRNA plus wild-type ArpC4, or ArpC4 shRNA plus 5KR mutant (each also expressing luciferase). Scale bar: 200 μm. (j) H1299 cells expressing either control or Flag-UFL1 were infected with lentiviruses expressing control shRNA, ArpC4 shRNA, ArpC4 shRNA plus wild-type ArpC4, or ArpC4 shRNA plus 5KR mutant as indicated. Cells lysates were subjected to western blot analysis. All experiments were repeated three times independently with similar results. The shown blots and images are representative of three independent experiments. Data in (a, c, g, h) are mean ± SD (n = 3). Data in (d–f) are mean ± SEM (n = 18, 14, 16 and 14, respectively). Statistical analysis was performed using one-way ANOVA (a, c, d, g, h).
Extended Data Fig. 6 K130 is crucial for mediating the functional effects of ArpC4 UFMylation on cell migration and invasion.
(a–g) A549 cells expressing ArpC4 shRNA were reconstituted with the indicated Flag-ArpC4 proteins. Cells were subjected to wound-healing (a and b), transwell migration (d and e), and transwell invasion assay (f and g). Cell lysates were analyzed by western blotting (c). Scale bar in a: 200 μm; in d and f: 100 μm. (h–j) H1299 cells expressing either control or Flag-UFL1 were infected with lentiviruses expressing control shRNA, ArpC4 shRNA, ArpC4 shRNA plus wild-type ArpC4, or ArpC4 shRNA plus ArpC4 mutants as indicated. Cells were subjected to transwell migration (h) and transwell invasion (j) assays. Cell lysates were analyzed by western blotting (i). Scale bars in h and j: 100 μm. (k) H1299 cells expressing control shRNA, UFL1 shRNA-#1, and UFL1 shRNA-#2 were serum-starved for 6 h before they were treated with or without 10% FBS for 30 min. Cell lysates were incubated with glutathione beads immobilized recombinant GST-PAK1-PBD. The input and GST-PAK1-PBD bound Rac1 were analyzed by western blotting. GST-PAK1-PBD bound Rac1 was considered as active Rac1. (l) Cell lysates from H1299 cells expressing control shRNA or UFL1 shRNA were immunoprecipitated with control IgG or anti-Arp2 antibody, followed by western blot analysis. The phosphorylation of Arp2 was detected using anti-phospho-Arp2 (T237/T238) antibody. (m) Cell lysates from H1299 cells expressing control shRNA or UFL1 shRNA were immunoprecipitated with control IgG or anti-ArpC1 antibody, followed by western blot analysis. The phosphorylation of ArpC1 was detected using pan phospho-serine/threonine antibody. All experiments were repeated three times independently with similar results. The shown blots and images are representative of three independent experiments. Data in (b, e, g, h, j) are mean ± SD (n = 3). Statistical analysis was performed using one-way ANOVA (b, e, g, h, j).
Extended Data Fig. 7 Akt phosphorylates UFL1 at T426.
(a) H1299 cells expressing control or UFL1-TurboID were serum-starved for 6 h before they were treated with or without 10% FBS for 30 min in the presence of 200 μM D-biotin. Biotinylated proteins were pulled-down using streptavidin beads (Thermo Fisher Scientific). The input and biotinylated proteins were analyzed by western blotting. (b) H1299 cells expressing control or Flag-UFL1 were starved for 6 h and treated with MK2206 or PD98059 for 2 h, followed by 10% FBS stimulation for 30 min. Cell lysates were subjected to immunoprecipitation with anti-Flag antibody and western blot analysis. (c) Prediction of potential phosphorylation sites on UFL1 using Group-based Prediction System (GPS). (d) Dot blot analysis of the generated phospho-UFL1-T426 antibody against the indicated UFL1 peptides with or without T426 phosphorylation, which demonstrated that the p-UFL1(T426) antibody specifically recognize the pT426 epitope. (e) Flag-tagged UFL1 alone or together with HA-Akt1 was expressed in HEK293T cells. The cells were treated with or without MK2206 for 2 h before they were subjected to immunoprecipitation with anti-Flag antibody, followed by western blot analysis. (f) Recombinant GST-UFL1 or UFL1-T426A were incubated with purified Flag-Akt1 in the kinase buffer for 30 min at 30 °C. The phosphorylation of UFL1 was detected by western blot using anti-p-UFL1 (T426) antibody. All experiments were repeated three times independently with similar results. The shown blots are representative of three independent experiments.
Extended Data Fig. 8 T426 phosphorylation is essential for UFL1 to promote cell migration, invasion, and metastasis.
(a–c) UFL1 knockdown A549 cells were reconstituted with the indicated Flag-UFL1 proteins. Cells were subjected to wound-healing (a) and transwell migration (c) assay. Cell lysates were also analyzed by western blot (b). Scale bar in a: 200 μm; c: 100 μm. (d–f) The analysis of cell migration speed (d), persistence (e), and mean-squared displacement (MSD) (f) of Fig. 8d. (g) UFL1 knockdown A549 cells were reconstituted with the indicated Flag-UFL1 proteins. Cells were subjected transwell invasion assay. Scale bar: 100 μm; (h) The percentage of lamellipodia-positive cells in Fig. 8f. (i, j) UFL1 knockdown A549 cells were reconstituted with the indicated Flag-UFL1 proteins. Cells were serum-starved for 6 h before they were scratched. Cells were then treated with 10% FBS for 30 min, followed by immunofluorescence assay. (i) The lamellipodia protrusions driven by branched actin networks were shown by immunostaining with anti-Cortactin antibody. White arrows indicate lamellipodia region. (j) The percentage of lamellipodia-positive cells were calculated. Scale bar: 20 μm. (k) H&E staining of lung lobes from the NCG mice intravenously injected with A549 cells expressing control shRNA, UFL1 shRNA, UFL1 shRNA plus wild-type UFL1, UFL shRNA plus UFL1-T426A, or UFL shRNA plus UFL1-T426D (each also expressing luciferase). Scale bar: 200 μm. All experiments were repeated three times independently with similar results. The shown blots and images are representative of three independent experiments. Data in (a, c, g, h, j) are mean ± SD (n = 3). Data in (d–f) are mean ± SEM (n = 15 for each group). Statistical analysis was performed using one-way ANOVA (a, c, d, g, h, j).
Extended Data Fig. 9 Analysis of UFL1 phosphorylation in clinical lung cancer samples.
(a, b) UFL1 and phospho-UFL1 (T426) levels in lung cancer tissues (T) and matched adjacent normal tissues (N) (a). The expression levels of UFL1 in (a) and (Fig. 8i) were quantified and normalized to β-actin (b). (c, d) Levels of UFL1, phospho-UFL1 (T426), and phospho-Akt (S473) in lung cancer tissues (c). UFL1 levels in (c) and (Fig. 8k) were quantified and normalized to β-actin (d). All experiments were repeated three times independently with similar results. The shown blots are representative of three independent experiments. Data in (b) were analyzed by a paired Student’s t test (two-tailed) (n = 33). Data in (d) are mean ± SD (n = 22, 13 and 11, respectively), analyzed by one-way ANOVA.
Supplementary information
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 7
Unprocessed western blots.
Source Data Fig. 8
Unprocessed western blots.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Unprocessed western blots.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, K., Hu, H., Fang, D. et al. Akt-phosphorylated UFL1 UFMylates ArpC4 to promote metastasis. Nat Struct Mol Biol 32, 1528–1541 (2025). https://doi.org/10.1038/s41594-025-01576-8
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41594-025-01576-8
This article is cited by
-
UFMylation of ARPC4 facilitates lamellipodia formation and promotes cancer metastasis
Nature Structural & Molecular Biology (2025)