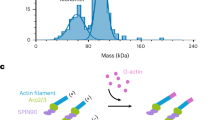
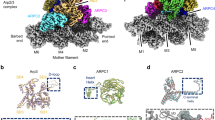
Abstract
Arp2/3 complex is a key nucleator of actin filaments. It requires activation by nucleation-promoting factors (NPFs). WISH/DIP1/SPIN90 (WDS) proteins represent a unique class of NPFs that activate the Arp2/3 complex independently of preexisting filaments, promoting linear actin-filament nucleation. In fission yeast, Dip1 binds to the clamp subunits in Arp2/3 complex to induce the short-pitch conformation, where Arp2 moves closer to Arp3 to mimic a filamentous actin dimer. However, how WDS proteins stimulate subunit flattening in Arp subunits, a ‘scissor-like’ conformational change akin to what is observed in an actin monomer during filament formation, remained unclear. Here we present cryo-electron microscopy structures of human SPIN90 bound to activated bovine Arp2/3 complex on an actin filament pointed end. The structures show that SPIN90 dimerizes through a metazoan-specific domain in the middle segment, engaging both the clamp and the Arp3/ARPC3 interface, to drive the activating conformational changes in Arp2/3 complex. Remarkably, a single SPIN90 dimer can also bridge two Arp2/3 complexes, enabling bidirectional actin nucleation and suggesting a mechanism for rapidly assembling complex actin network architectures.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
EM maps of the singlet and doublet SPIN90–Arp2/3 complex-nucleated filament assemblies were deposited to the EM Data Bank under accession codes EMD-63642 and EMD-63658, respectively. The corresponding atomic models were deposited to the PDB under accession codes 9M5E and 9M64, respectively. The individual focused maps for the Arp2/3 complex, SPIN90 dimer and actin-filament subregions for the singlet complex were deposited to the EM Data Bank under accession codes EMD-63880, EMD-63883 and EMD-63886, respectively. For the doublet complex, individual focused maps for the two Arp2/3 complexes, SPIN90 dimer and two actin-filament subregions were deposited to the EM Data Bank under accession codes EMD-63887, EMD-63888, EMD-63889, EMD-63890 and EMD-63891, respectively. Data and materials can be obtained from the corresponding authors upon request. Source data are provided with this paper.
References
Pollard, T. D. & Cooper, J. A. Actin, a central player in cell shape and movement. Science 326, 1208–1212 (2009).
Rosenbloom, A. D., Kovar, E. W., Kovar, D. R., Loew, L. M. & Pollard, T. D. Mechanism of actin filament nucleation. Biophys. J. 120, 4399–4417 (2021).
Chesarone, M. A. & Goode, B. L. Actin nucleation and elongation factors: mechanisms and interplay. Curr. Opin. Cell Biol. 21, 28–37 (2009).
Skau, C. T. & Waterman, C. M. Specification of architecture and function of actin structures by actin nucleation factors. Annu. Rev. Biophys. 44, 285–310 (2015).
Rotty, J. D., Wu, C. & Bear, J. E. New insights into the regulation and cellular functions of the Arp2/3 complex. Nat. Rev. Mol. Cell Biol. 14, 7–12 (2013).
Narvaez-Ortiz, H. Y. & Nolen, B. J. Unconcerted conformational changes in Arp2/3 complex integrate multiple activating signals to assemble functional actin networks. Curr. Biol. 32, 975–987 (2022).
Zimmet, A. et al. Cryo-EM structure of NPF-bound human Arp2/3 complex and activation mechanism. Sci. Adv. 6, 7651–7656 (2020).
Mullins, R. D., Bieling, P. & Fletcher, D. A. From solution to surface to filament: actin flux into branched networks. Biophys. Rev. 10, 1537–1551 (2018).
Rodnick-Smith, M., Luan, Q., Liu, S.-L. & Nolen, B. J. Role and structural mechanism of WASP-triggered conformational changes in branched actin filament nucleation by Arp2/3 complex. Proc. Natl Acad. Sci. USA 113, E3834–E3843 (2016).
Espinoza-Sanchez, S., Metskas, L. A., Chou, S. Z., Rhoades, E. & Pollard, T. D. Conformational changes in Arp2/3 complex induced by ATP, WASp-VCA, and actin filaments. Proc. Natl Acad. Sci. USA 115, E8642–E8651 (2018).
Xu, X.-P. et al. Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 31, 236–247 (2012).
Fäßler, F., Dimchev, G., Hodirnau, V.-V., Wan, W. & Schur, F. K. M. Cryo-electron tomography structure of Arp2/3 complex in cells reveals new insights into the branch junction. Nat. Commun. 11, 6437 (2020).
Saks, A. J., Barrie, K. R., Rebowski, G. & Dominguez, R. NPF binding to Arp2 is allosterically linked to the release of ArpC5’s N-terminal tail and conformational changes in Arp2/3 complex. Proc. Natl Acad. Sci. USA 122, e2421557122 (2025).
Chou, S. Z., Chatterjee, M. & Pollard, T. D. Mechanism of actin filament branch formation by Arp2/3 complex revealed by a high-resolution cryo-EM structure of the branch junction. Proc. Natl Acad. Sci. USA 119, e2206722119 (2022).
Liu, T. et al. Cortactin stabilizes actin branches by bridging activated Arp2/3 to its nucleated actin filament. Nat. Struct. Mol. Biol. 31, 801–809 (2024).
Ding, B. et al. Structure of Arp2/3 complex at a branched actin filament junction resolved by single-particle cryo-electron microscopy. Proc. Natl Acad. Sci. USA 119, e2202723119 (2022).
Shaaban, M., Chowdhury, S. & Nolen, B. J. Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state. Nat. Struct. Mol. Biol. 27, 1009–1016 (2020).
Chavali, S. S. et al. Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex. Nat. Commun. 15, 2059 (2024).
Wagner, A. R., Luan, Q., Liu, S. L. & Nolen, B. J. DIP1 defines a class of Arp2/3 complex activators that function without preformed actin filaments. Curr. Biol. 23, 1990–1998 (2013).
Liu, S. L. et al. Analysis of functional surfaces on the actin nucleation promoting factor DIP1 required for Arp2/3 complex activation and endocytic actin network assembly. J. Biol. Chem. 298, 102019 (2022).
Dae, J. K. et al. Interaction of SPIN90 with the Arp2/3 complex mediates lamellipodia and actin comet tail formation. J. Biol. Chem. 281, 617–625 (2006).
Luan, Q., Liu, S., Helgeson, L. A. & Nolen, B. J.Structure of the nucleation‐promoting factor SPIN 90 bound to the actin filament nucleator Arp2/3 complex. EMBO J. 37, e100005 (2018).
Balzer, C. J., Wagner, A. R., Helgeson, L. A. & Nolen, B. J. Single-turnover activation of Arp2/3 complex by DIP1 may balance nucleation of linear versus branched actin filaments. Curr. Biol. 29, 3331–3338 (2019).
Achard, V. et al. A ‘primer’-based mechanism underlies branched actin filament network formation and motility. Curr. Biol. 20, 423–428 (2010).
Machesky, L. M. et al. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl Acad. Sci. USA 96, 3739–3744 (1999).
Kim, H. et al. SPIN90, an adaptor protein, alters the proximity between Rab5 and Gapex5 and facilitates Rab5 activation during EGF endocytosis. Exp. Mol. Med. 51, 1–14 (2019).
Basu, R. & Chang, F. Characterization of DIP1p reveals a switch in Arp2/3-dependent actin assembly for fission yeast endocytosis. Curr. Biol. 21, 905–916 (2011).
Cao, L. et al. SPIN90 associates with mDia1 and the Arp2/3 complex to regulate cortical actin organization. Nat. Cell Biol. 22, 803–814 (2020).
Luan, Q., Zelter, A., MacCoss, M. J., Davis, T. N. & Nolen, B. J. Identification of Wiskott–Aldrich syndrome protein (WASP) binding sites on the branched actin filament nucleator Arp2/3 complex. Proc. Natl Acad. Sci. USA 115, E1409–E1418 (2018).
Lim, C. S., Kim, S. H., Jung, J. G., Kim, J. K. & Song, W. K. Regulation of SPIN90 phosphorylation and interaction with Nck by ERK and cell adhesion. J. Biol. Chem. 278, 52116–52123 (2003).
Lim, C. S. et al. SPIN90 (SH3 protein interacting with Nck, 90 kDa), an adaptor protein that is developmentally regulated during cardiac myocyte differentiation. J. Biol. Chem. 276, 12871–12878 (2001).
Cao, L., Ghasemi, F., Way, M., Jégou, A. & Romet-Lemonne, G. Regulation of branched versus linear Arp2/3-generated actin filaments. EMBO J. 42, e113008 (2023).
Reynolds, M. J., Hachicho, C., Carl, A. G., Gong, R. & Alushin, G. M. Bending forces and nucleotide state jointly regulate F-actin structure. Nature 611, 380–386 (2022).
Liu, T. et al. Arp2/3-mediated bidirectional actin assembly by SPIN90 dimers in metazoans. Preprint at bioRxiv https://doi.org/10.1101/2025.01.31.635869 (2025).
Nolen, B. J. & Pollard, T. D. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol. Cell 26, 449–457 (2007).
Punjani, A. & Fleet, D. J. 3D variability analysis: resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021).
Ti, S.-C., Jurgenson, C. T., Nolen, B. J. & Pollard, T. D. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc. Natl Acad. Sci. USA 108, E463–E471 (2011).
Padrick, S. B., Doolittle, L. K., Brautigam, C. A., King, D. S. & Rosen, M. K.Arp2/3 complex is bound and activated by two WASP proteins. Proc. Natl Acad. Sci. USA 108, E472–E479 (2011).
Balzer, C. J. et al. Synergy between Wsp1 and DIP1 may initiate assembly of endocytic actin networks. eLife 9, e60419 (2020).
Singh, Y., Hocky, G. M. & Nolen, B. J. Molecular dynamics simulations support a multistep pathway for activation of branched actin filament nucleation by Arp2/3 complex. J. Biol. Chem. 299, 105169 (2023).
Ingerman, E., Hsiao, J. Y. & Mullins, R. D. Arp2/3 complex ATP hydrolysis promotes lamellipodial actin network disassembly but is dispensable for assembly. J. Cell Biol. 200, 619–633 (2013).
Pardee, J. D. & Spudich, J. A.Purification of muscle actin. Methods Enzymol. 85 Pt B, 164–181 (1982).
Ulrichs, H., Gaska, I. & Shekhar, S. Multicomponent regulation of actin barbed end assembly by twinfilin, formin and capping protein. Nat. Commun. 14, 3981 (2023).
Doolittle, L. K., Rosen, M. K. & Padrick, S. B. Purification of native Arp2/3 complex from bovine thymus. Methods Mol. Biol. 1046, 231–250 (2013).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D 74, 519–530 (2018).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in PHENIX. Acta Crystallogr. D 75, 861–877 (2019).
Williams, C. J. et al. MolProbity: more and better reference data for improved all‐atom structure validation. Protein Sci. 27, 293–315 (2018).
Berman, H., Henrick, K. & Nakamura, H. Announcing the worldwide Protein Data Bank. Nat. Struct. Mol. Biol. 10, 980–980 (2003).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Yariv, B. et al. Using evolutionary data to make sense of macromolecules with a ‘face-lifted’ ConSurf. Protein Sci. 32, e4582 (2023).
Drenckhahn, D. & Pollard, T. D. Elongation of actin filaments is a diffusion-limited reaction at the barbed end and is accelerated by inert macromolecules. J. Biol. Chem. 261, 12754–12758 (1986).
Acknowledgements
Cryo-EM data were collected at the Council of Scientific and Industrial Research Center for Cellular and Molecular Biology (CSIR-CCMB) EM Facility. We gratefully acknowledge H. Adicherla, S. Shrivastava and R. Reddi for their valuable support and assistance at this facility. Computational resources and support were provided by the high-performance computing facilities at CSIR-CCMB and we extend our thanks to G. Thanu, A. Kumari and K. R. Chary for their help and assistance. We also express our gratitude to the director of CSIR-CCMB for his generous support. We also thank K. Garai for providing access to the SEC–MALS instrument at the Tata Institute of Fundamental Research and D. Saraswati for her valuable assistance with the SEC–MALS experiments and analyses. This research was funded by CSIR grants FBR070301 and OLP0028 to S.C. and National Institutes of Health grant 5R35GM136319 to B.J.N. The CSIR-CCMB EM Facility was established with support from CSIR grant FAC008. J.F. and A.K.P. are supported by senior research fellowships from CSIR and K.V.E. is supported by a junior research fellowship from the University Grants Commission.
Author information
Authors and Affiliations
Contributions
S.C., J.F. and B.J.N. conceptualized the project and prepared the paper with input from all authors. J.F. and S.C. determined the biochemical conditions for sample preparation. Cryo-EM grid vitrification, data collection and processing were carried out by J.F. J.F. and A.K.P. built the atomic models. Structural analysis was performed by J.F. and S.C. Protein purification and biochemical assays were performed by K.T.S., S.S., R.M., J.F. and H.Y.N.-O. Sequence analysis was performed by K.V.E. The project was supervised in its entirety by S.C.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Katarzyna Ciazynska, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Components purified for reconstituting complexes, and cryo-EM data.
a, Coomassie stained SDS-PAGE gel of proteins used for the structural studies and for pyrene actin polymerization assays. b, A representative micrograph out of a dataset comprising of 24,379 micrographs highlighting the two complexes: Singlets circled in orange and doublets in green, respectively. Scale bar: 100 nm. Different views of the initial 2D class averages of the singlet complex (highlighted in orange) and doublet complex (highlighted in green) are shown on the right. These were used as initial templates for particle picking from micrographs.
Extended Data Fig. 2 SPIN90 dimerization is mediated by the MSDR.
a, Comparison of the Buried Surface Area (BSA) between symmetry-related SPIN90 molecules in the crystal structure (PDB:6DEE) with the singlet and doublet complexes. Also highlighted are the RMSD values between the SPIN90 dimer and individual molecules in the singlet and doublet complexes respectively. b, The N-terminal eighteen residues containing the first helix of the MSDR, that is truncated in the 326-722 construct of SPIN90, is highlighted (with the rest of the MSDR faded). c, Size-exclusion chromatography coupled with multi-angle light scattering (SEC-MALS) profiles are shown for two SPIN90 constructs (left-side panel): one containing the full MSDR region (SPIN90 (269-722); pink chromatogram trace, peak labeled ‘I’) another lacking the N-terminal eighteen residues of the MSDR (SPIN90 (326-722); magenta chromatogram trace, peak labeled ‘II’). The right-side panels show MALS-derived molecular weight analyses of the peak regions. The measured molecular weight of SPIN90 (269-722) construct is ~100 ± 20 kDa (theoretical molecular mass from sequence: 50.3 kDa), consistent with a dimeric species. In contrast, SPIN90 (326-722) (∆MSDR) construct shows a molecular weight of ~50 ± 5 kDa (theoretical molecular mass from sequence: 46.9 kDa), indicating it exists predominantly as a monomer in solution. These data suggest that the N-terminal region of the MSDR is critical for dimerization. d, Comparison between B-factors of SPIN90 dimers in the singlet and doublet complexes shows regions in the singlet SPIN90 dimer with higher flexibility compared to the doublet which is stabilized by the other apposing Arp2/3 complex.
Extended Data Fig. 3 Overall data processing workflow used for 3D reconstruction of the complexes.
After iterative rounds of template picking and 2D classification, workflow used for 3D reconstruction of the singlet complex is shown (left side of the gray dashed line), with the reconstructed volume colored by local resolution, 3D FSC plot, and map-to-model FSC plot shown for map quality and resolution assessment (at the bottom left of the workflow) for the same. Similarly, the right side of the image shows the workflow for the doublet complex with its corresponding reconstructed volume colored by local resolution, 3D FSC plot, and map-to-model FSC plot (at the bottom right).
Extended Data Fig. 5 Commonalities and differences between WDS proteins of protozoans and metazoans and their effects on cross-species Arp2/3 complex activity.
a, Conserved leucine rich region within the ARM domain in SPIN90 and Dip1 binds to the clamp subunits (blue and cyan) of Arp2/3 complex. The mother filament of actin also makes extensive contact with the clamp subunits in Arp2/3 complex. b, Plot of the clamp dihedrals versus the distances between the centers of geometry in Arp2 and Arp3. SPIN90 activated structures show Arp2-Arp3 short pitch distance is closest to the short pitch distance in actin filament. c, Time course of pyrene actin polymerization for reactions containing 3 μM 15% pyrene actin, 50 nM of Bos taurus (Bt) Arp2/3 complex (left panel), or 50 nM of S. pombe (Sp) Arp2/3 complex (right panel), and SPIN90 or Dip1, as indicated. These show that protozoan WDS protein fails to activate metazoan Arp2/3 complex and vice versa.
Extended Data Fig. 6 Conservation of SPIN90 residues involved in dimerization and interactions with Arp2/3 complex.
a, Highly conserved residues of SPIN90 shown in purple belong to regions (1) involved in dimerization of SPIN90 (MSDR) in MS, (2) interaction with Arp3, and (3) clamp binding via the leucine rich domain (LRD) based on ConSurf analysis. b, Multiple sequence alignment of human SPIN90 with ten other metazoan species. Residues that interact with Arp2/3 complex and show sequence identity across metazoans have been highlighted. Details of sequences and alignments have been described in the methods.
Extended Data Fig. 7 Flattening of Arps and SPIN90’s interaction with Arp3.
a, Overlay (overlayed on SD3 and SD4) of twisted Arp3 (white colored, from inactive Arp2/3 complex (PDB: 4JD2)) on flattened Arp3 (orange, active Arp2/3 complex) in the left side of the image, and similarly for Arp2 (twisted in white, and flattened in red) is shown on the right side of the image. The dihedrals between the subdomains are shown as green bonds connecting the centre of masses of each subdomain (shown as green spheres). The direction of flattening during activation is shown by the curved black arrows for both the Arps. b, SPIN90’s insertion loop does not make any contact with Arp3 in the inactive complex, compared to the insertion of this loop into Arp3’s pointed end cleft in the active complex. The dashed bold yellow lines indicate the separation distances between the Cα backbone atoms for W401 of SPIN90 and R197 of Arp3 respectively c, Reconstructed density between W401 of SPIN90 and R198 of Arp3 emphasizing the cation-pie interaction. d, The SPIN90 insertion loop penetrates more deeply into the Arp3 pointed end cleft than the loop from the filamentous actin subunit in the branch junction (PDB: 7TPT). The dashed box highlighting the distance between Cα backbone atom of D172 of Arp3 to N397 of SPIN90 in the SPIN90-Arp2/3 complex activated structure (left side), and N360 of actin subunit from the mother filament in the branched actin junction structure (PDB: 7TPT) (right side).
Supplementary information
Supplementary Video 1
Flexibility of the doublet complex revealed by 3DVAs.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Uncropped SDS gel image.
Source Data Extended Data Fig. 2
SEC–MALS statistical source data.
Source Data Extended Data Fig. 5
Source data for the distances between the centers of geometry in Arp2 and Arp3 and statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Francis, J., Pathri, A.K., Shyam, K.T. et al. Activation of Arp2/3 complex by a SPIN90 dimer in linear actin-filament nucleation. Nat Struct Mol Biol 32, 2272–2284 (2025). https://doi.org/10.1038/s41594-025-01673-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41594-025-01673-8