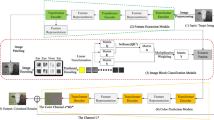
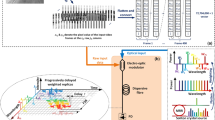
Abstract
Recently, there has been an explosion of scientific literature describing the use of colorimetry for monitoring the progression or the endpoint result of colorimetric reactions. The availability of inexpensive imaging technology (e.g., scanners, Raspberry Pi, smartphones and other sub-$50 digital cameras) has lowered the barrier to accessing cost-efficient, objective detection methodologies. However, to exploit these imaging devices as low-cost colorimetric detectors, it is paramount that they interface with flexible software that is capable of image segmentation and probing a variety of color spaces (RGB, HSB, Y’UV, L*a*b*, etc.). Development of tailor-made software (e.g., smartphone applications) for advanced image analysis requires complex, custom-written processing algorithms, advanced computer programming knowledge and/or expertise in physics, mathematics, pattern recognition and computer vision and learning. Freeware programs, such as ImageJ, offer an alternative, affordable path to robust image analysis. Here we describe a protocol that uses the ImageJ program to process images of colorimetric experiments. In practice, this protocol consists of three distinct workflow options. This protocol is accessible to uninitiated users with little experience in image processing or color science and does not require fluorescence signals, expensive imaging equipment or custom-written algorithms. We anticipate that total analysis time per region of interest is ~6 min for new users and <3 min for experienced users, although initial color threshold determination might take longer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
Data availability
All figures and any associated data have not been previously published. The source image files used to generate the primary data underlying the figures featured in this protocol are available via open access at zenodo.org, which can be found at https://doi.org/10.5281/zenodo.3976070.
Code availability
This protocol requires no custom scripts or algorithms. Rather, this protocol uses free, publicly available software packages (e.g., ImageJ 2.0.0-rc-69/1.52p bundled with Java 1.8.0_172 (32-bit) (https://imagej.nih.gov/ij/download.html) and the Fiji distribution of ImageJ (https://imagej.net/Fiji/Downloads))10,11,12 as well as available commercial software (e.g., Microsoft Excel for Microsoft 365).
References
Lee, B. B. The evolution of concepts of color vision. Neurociencias 4, 209–224 (2008).
Molday, R. S. & Moritz, O. L. Photoreceptors at a glance. J. Cell Sci. 128, 4039–4045 (2015).
Jameson, K. A. in The Oxford Companion to Consciousness 155–158 (Oxford University Press, 2009).
Krauss, S. T. et al. Objective method for presumptive field-testing of illicit drug possession using centrifugal microdevices and smartphone analysis. Anal. Chem. 88, 8689–8697 (2016).
CIE. Commission internationale de l’eclairage proceedings, 1931 (Cambridge University, 1932).
Smith, T. & Guild, J. The CIE colorimetric standards and their use. Trans. Opt. Soc. 33, 73 (1931).
CIE. Colorimetry-Part 4: CIE 1976 L* a* b* colour space. International Organization for Standardization https://www.iso.org/standard/74166.html (2008).
Schnapf, J., Kraft, T. & Baylor, D. Spectral sensitivity of human cone photoreceptors. Nature 325, 439–441 (1987).
Anderson, M., Motta, R., Chandrasekar, S. & Stokes, M. in Color and Imaging Conference 238–245 (Society for Imaging Science and Technology, 1996).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Rueden, C. T. et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676 (2012).
Roels, J. et al. An interactive ImageJ plugin for semi-automated image denoising in electron microscopy. Nat. Commun. 11, 1–13 (2020).
Boudaoud, A. et al. FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nat. Protoc. 9, 457 (2014).
Krauss, S. T., Holt, V. C. & Landers, J. P. Simple reagent storage in polyester-paper hybrid microdevices for colorimetric detection. Sens. Actuators B Chem. 246, 740–747 (2017).
Krauss, S. T., Nauman, A. Q., Garner, G. T. & Landers, J. P. Color manipulation through microchip tinting for colorimetric detection using hue image analysis. Lab Chip 17, 4089–4096 (2017).
Krauss, S. T. et al. Centrifugal microfluidic devices using low-volume reagent storage and inward fluid displacement for presumptive drug detection. Sens. Actuators B Chem. 284, 704–710 (2019).
Thompson, B. L., Wyckoff, S. L., Haverstick, D. M. & Landers, J. P. Simple, reagentless quantification of total bilirubin in blood via microfluidic phototreatment and image analysis. Anal. Chem. 89, 3228–3234 (2017).
Jackson, K. R. et al. A novel loop-mediated isothermal amplification method for identification of four body fluids with smartphone detection. Forensic Sci. Int. Genet. 45, 102195 (2020).
Russell, R. A. et al. Segmentation of fluorescence microscopy images for quantitative analysis of cell nuclear architecture. Biophys. J. 96, 3379–3389 (2009).
Balsam, J., Bruck, H. A., Kostov, Y. & Rasooly, A. Image stacking approach to increase sensitivity of fluorescence detection using a low cost complementary metal-oxide-semiconductor (CMOS) webcam. Sens. Actuators B Chem. 171, 141–147 (2012).
Perez, A. J. et al. A workflow for the automatic segmentation of organelles in electron microscopy image stacks. Front. Neuroanat. 8, 126 (2014).
Stegmaier, J. et al. Fast segmentation of stained nuclei in terabyte-scale, time resolved 3D microscopy image stacks. PLoS ONE 9, e90036 (2014).
Capitan-Vallvey, L. F., Lopez-Ruiz, N., Martinez-Olmos, A., Erenas, M. M. & Palma, A. J. Recent developments in computer vision-based analytical chemistry: a tutorial review. Anal. Chim. Acta 899, 23–56 (2015).
Cao, S., Huang, D., Wang, Y. & Li, G. in Advances in Mechanical and Electronic Engineering 381–386 (Springer, 2012).
Cantrell, K., Erenas, M., de Orbe-Payá, I. & Capitán-Vallvey, L. Use of the hue parameter of the hue, saturation, value color space as a quantitative analytical parameter for bitonal optical sensors. Anal. Chem. 82, 531–542 (2010).
Ibraheem, N. A., Hasan, M. M., Khan, R. Z. & Mishra, P. K. Understanding color models: a review. ARPN J. Sci. Technol. 2, 265–275 (2012).
Hanbury, A. The taming of the hue, saturation and brightness colour space. Proc. 7th Computer Vision Winter Workshop http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.3.2574 (2002).
Barthel, K. U. 3D-data representation with ImageJ. Proc. 1st ImageJ User & Developer Conference http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.414.725 (2006).
Clark, C. P. et al. Closable valves and channels for polymeric microfluidic devices. Micromachines 11, 627 (2020).
Tian, Z., Liu, L. Q., Peng, C., Chen, Z. & Xu, C. A new development of measurement of 19-Nortestosterone by combining immunochromatographic strip assay and ImageJ software. Food Agric. Immunol. 20, 1–10 (2009).
Hwang, J., Kwon, D., Lee, S. & Jeon, S. Detection of Salmonella bacteria in milk using gold-coated magnetic nanoparticle clusters and lateral flow filters. RSC Adv. 6, 48445–48448 (2016).
Kortli, S. et al. Yersinia pestis detection using biotinylated dNTPs for signal enhancement in lateral flow assays. Anal. Chim. Acta 1112, 54–61 (2020).
Adkins, J. A. et al. Colorimetric and electrochemical bacteria detection using printed paper-and transparency-based analytic devices. Anal. Chem. 89, 3613–3621 (2017).
Tanner, N. A., Zhang, Y. & Evans, T. C. Jr Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques 58, 59–68 (2015).
Zhang, Y. et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. Preprint at https://www.medrxiv.org/content/10.1101/2020.02.26.20028373v1 (2020).
Singh, H. K., Tomar, S. K. & Maurya, P. K. Thresholding techniques applied for segmentation of RGB and multispectral images. https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.735.9363&rep=rep1&type=pdf (2012).
Acknowledgements
We gratefully acknowledge K. Siller (UVA Research Computing, University of Virginia) for his advice and instruction provided in ImageJ-Fiji workshops given at the University of Virginia (attended by M.S.W. and A.T.S. in 2018). We also recognize the initial foundational image analysis work of previous Landers lab members, including C. P. Clark, S. T. Krauss, K. R. Jackson, J. Li, D. A. Nelson, O. N. Scott and B. L. Thompson.
Author information
Authors and Affiliations
Contributions
All author contributions are based on CRediT standards. Writing—original draft: M.S.W. Writing—review and editing: all authors. Conceptualization, methodology, investigation, data curation, formal analysis and visualization of the image segmentation procedure: M.S.W. Conceptualization, methodology, investigation and image acquisition for all colorimetric experiments: M.S.W., L.M.D. and A.T.S. Project administration and funding acquisition: J.P.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Bahram Hemmateenejad and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Clark, C. P. et al. Micromachines 11, 627 (2020): https://doi.org/10.3390/mi11070627
Jackson, K. R. et al. Forensic Sci. Int. Genet. 45, 102195 (2020): https://doi.org/10.1016/j.fsigen.2019.102195
Krauss, S. T. et al. Sens. Actuators B Chem. 284, 704−710 (2019): https://doi.org/10.1016/j.snb.2018.12.113
Krauss, S.T. et al. Lab Chip 17, 4089−4096 (2017): https://doi.org/10.1039/C7LC00796E
Rights and permissions
About this article
Cite this article
Woolf, M.S., Dignan, L.M., Scott, A.T. et al. Digital postprocessing and image segmentation for objective analysis of colorimetric reactions. Nat Protoc 16, 218–238 (2021). https://doi.org/10.1038/s41596-020-00413-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-020-00413-0
This article is cited by
-
The underperforming Abbott-Bioline Malaria Ag P.f/P.v rapid diagnostic test: a whiter shade of pale - where the truth is not plain to see
Malaria Journal (2025)
-
Rapid antimicrobial susceptibility tests performed by self-diluting microfluidic chips for drug resistance studies and point-of-care diagnostics
Microsystems & Nanoengineering (2025)
-
Pre- and Postoperative Visualization in Rhinoplasty: Effects of LED Light Panels in Aesthetic Photography on Image Integrity
Aesthetic Plastic Surgery (2025)
-
MSI-based damage detection and damage degree classification study of snow pear
Agricultural Products Processing and Storage (2025)
-
GelMA loaded with platelet lysate promotes skin regeneration and angiogenesis in pressure ulcers by activating STAT3
Scientific Reports (2024)