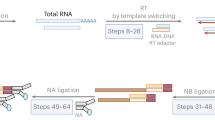
Abstract
Elongation, splicing and polyadenylation are fundamental steps of transcription, and studying their coordination requires simultaneous monitoring of these dynamic processes on one transcript. We recently developed a full-length nascent RNA sequencing method in the model plant Arabidopsis that simultaneously detects RNA polymerase II position, splicing status, polyadenylation site and poly(A) tail length at genome-wide scale. This method allows calculation of the kinetics of cotranscriptional splicing and detects polyadenylated transcripts with unspliced introns retained at specific positions posttranscriptionally. Here we describe a detailed protocol for this method called FLEP-seq (full-length elongating and polyadenylated RNA sequencing) that is applicable to plants. Library production requires as little as one nanogram of nascent RNA (after rRNA/tRNA removal), and either Nanopore or PacBio platforms can be used for sequencing. We also provide a complete bioinformatic pipeline from raw data processing to downstream analysis. The minimum time required for FLEP-seq, including RNA extraction and library preparation, is 36 h. The subsequent long-read sequencing and initial data analysis ranges between 31 and 40 h, depending on the sequencing platform.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
The data used in this study were previously published35 and deposited at NCBI under the accession number PRJNA591665.
Code availability
All software used in this study is described in detail in the Equipment section. Custom Python and R code used for bioinformatics analysis have been deposited in a GitHub repository (https://github.com/ZhaiLab-SUSTech/FLEPSeq).
References
Herzel, L., Ottoz, D. S. M., Alpert, T. & Neugebauer, K. M. Splicing and transcription touch base: co-transcriptional spliceosome assembly and function. Nat. Rev. Mol. Cell Biol. 18, 637–650 (2017).
Naftelberg, S., Schor, I. E., Ast, G. & Kornblihtt, A. R. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 84, 165–198 (2015).
Bentley, D. L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15, 163–175 (2014).
Carrillo Oesterreich, F. et al. Splicing of nascent RNA coincides with intron exit from RNA Polymerase II. Cell 165, 372–381 (2016).
Herzel, L., Straube, K. & Neugebauer, K. M. Long-read sequencing of nascent RNA reveals coupling among RNA processing events. Genome Res. 28, 1008–1019 (2018).
Reimer, K. A., Mimoso, C. A., Adelman, K. & Neugebauer, K. M. Co-transcriptional splicing regulates 3’ end cleavage during mammalian erythropoiesis. Mol. Cell https://doi.org/10.1016/j.molcel.2020.12.018 (2021).
Drexler, H. L., Choquet, K. & Churchman, L. S. Splicing kinetics and coordination revealed by direct nascent RNA sequencing through nanopores. Mol. Cell 77, 985–998.e988 (2020).
Drexler, H. L. et al. Revealing nascent RNA processing dynamics with nano-COP. Nat. Protoc. https://doi.org/10.1038/s41596-020-00469-y (2021).
Sousa-Luis, R. et al. POINT technology illuminates the processing of polymerase-associated intact nascent transcripts. Mol. Cell 81, 1935–1950.e6 (2021).
Wissink, E. M., Vihervaara, A., Tippens, N. D. & Lis, J. T. Nascent RNA analyses: tracking transcription and its regulation. Nat. Rev. Genet. 20, 705–723 (2019).
Mayer, A. et al. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161, 541–554 (2015).
Bhatt, DevM. et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150, 279–290 (2012).
Nechaev, S. et al. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327, 335 (2010).
Churchman, L. S. & Weissman, J. S. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469, 368–373 (2011).
Nojima, T. et al. Mammalian NET-Seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 161, 526–540 (2015).
Core, L. J., Waterfall, J. J. & Lis, J. T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845 (2008).
Kwak, H., Fuda, N. J., Core, L. J. & Lis, J. T. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953 (2013).
Judd, J. et al. A rapid, sensitive, scalable method for Precision Run-On sequencing (PRO-seq). Preprint at bioRxiv https://doi.org/10.1101/2020.05.18.102277 (2020).
Rabani, M. et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 29, 436–442 (2011).
Schwalb, B. et al. TT-seq maps the human transient transcriptome. Science 352, 1225 (2016).
Herzog, V. A. et al. Thiol-linked alkylation of RNA to assess expression dynamics. Nat. Methods 14, 1198–1204 (2017).
Schofield, J. A., Duffy, E. E., Kiefer, L., Sullivan, M. C. & Simon, M. D. TimeLapse-seq: adding a temporal dimension to RNA sequencing through nucleoside recoding. Nat. Methods 15, 221–225 (2018).
Shepard, P. J. et al. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17, 761–772 (2011).
Harrison, P. F. et al. PAT-seq: a method to study the integration of 3’-UTR dynamics with gene expression in the eukaryotic transcriptome. RNA 21, 1502–1510 (2015).
Ozsolak, F. et al. Direct RNA sequencing. Nature 461, 814–818 (2009).
Chang, H., Lim, J., Ha, M. & Kim, V. N. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol. Cell 53 (2014).
Lim, J., Lee, M., Son, A., Chang, H. & Kim, V. N. mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development. Genes Dev. 30, 1671–1682 (2016).
Subtelny, A. O., Eichhorn, S. W., Chen, G. R., Sive, H. & Bartel, D. P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71 (2014).
Legnini, I., Alles, J., Karaiskos, N., Ayoub, S. & Rajewsky, N. FLAM-seq: full-length mRNA sequencing reveals principles of poly(A) tail length control. Nat. Methods 16, 879–886 (2019).
Liu, Y., Nie, H., Liu, H. & Lu, F. Poly(A) inclusive RNA isoform sequencing (PAIso−seq) reveals wide-spread non-adenosine residues within RNA poly(A) tails. Nat. Commun. 10, 5292 (2019).
Parker, M. T. et al. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. eLife 9, e49658 (2020).
Workman, R. E. et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods 16, 1297–1305 (2019).
Roach, N. P. et al. The full-length transcriptome of C. elegans using direct RNA sequencing. Genome Res 30, 299–312 (2020).
Kim, D. et al. The architecture of SARS-CoV-2 transcriptome. Cell 181, 914–921.e910 (2020).
Jia, J. et al. Post-transcriptional splicing of nascent RNA contributes to widespread intron retention in plants. Nat. Plants 6, 780–788 (2020).
Boutz, P. L., Bhutkar, A. & Sharp, P. A. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 29, 63–80 (2015).
Mauger, O., Lemoine, F. & Scheiffele, P. Targeted intron retention and excision for rapid gene regulation in response to neuronal activity. Neuron 92, 1266–1278 (2016).
Naro, C. et al. An orchestrated intron retention program in meiosis controls timely usage of transcripts during germ cell differentiation. Dev. Cell 41, 82–93.e84 (2017).
Gahura, O. et al. Prp45 affects Prp22 partition in spliceosomal complexes and splicing efficiency of non-consensus substrates. J. Cell. Biochem. 106, 139–151 (2009).
Siatecka, M., Reyes, J. L. & Konarska, M. M. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 13, 1983–1993 (1999).
Zhu, D. et al. The features and regulation of co-transcriptional splicing in Arabidopsis. Mol. Plant 13, 278–294 (2020).
Krause, M. et al. tailfindr: alignment-free poly(A) length measurement for Oxford Nanopore RNA and DNA sequencing. RNA 25, 1229–1241 (2019).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
Cheng, C.-Y. et al. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89, 789–804 (2017).
Acknowledgements
We are grateful for the useful comments and edits suggested by the anonymous reviewers. The group of J.Z. is supported by a Stable Support Plan Program of Shenzhen Natural Science Fund Grant (20200925153345004), a National Key R&D Program of China Grant (2019YFA0903903), the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2016ZT06S172), the Shenzhen Sci-Tech Fund (KYTDPT20181011104005) and the Key Laboratory of Molecular Design for Plant Cell Factory of Guangdong Higher Education Institutes (2019KSYS006).
Author information
Authors and Affiliations
Contributions
Y.L., J.J. and J.Z. conceived and designed the study. Y.L., J.J. and X.J. developed the method and performed the experiments. J.J., W.M. and J.Z. designed the computational pipeline and analyzed the data. J.Z. conceived and oversaw the study. All authors wrote and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Zongwei Cai, Shona Murphy, Yongsheng Shi and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Jia et al. Nat. Plants 6, 780–788 (2020): https://doi.org/10.1038/s41477-020-0688-1
Key data used in this protocol
Jia et al. Nat. Plants 6, 780–788 (2020): https://doi.org/10.1038/s41477-020-0688-1
Rights and permissions
About this article
Cite this article
Long, Y., Jia, J., Mo, W. et al. FLEP-seq: simultaneous detection of RNA polymerase II position, splicing status, polyadenylation site and poly(A) tail length at genome-wide scale by single-molecule nascent RNA sequencing. Nat Protoc 16, 4355–4381 (2021). https://doi.org/10.1038/s41596-021-00581-7
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41596-021-00581-7
This article is cited by
-
Nano3P-seq: charting the coding and noncoding transcriptome at single-molecule resolution
Nature Protocols (2025)
-
Pre-mRNA processing factors differentially impact coordination between co-transcriptional cleavage and transcription termination
Nature Communications (2025)
-
Profiling the epigenome using long-read sequencing
Nature Genetics (2025)
-
Transcriptomics in the era of long-read sequencing
Nature Reviews Genetics (2025)
-
Epigenetics in the modern era of crop improvements
Science China Life Sciences (2025)