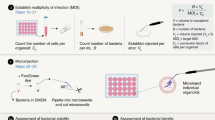
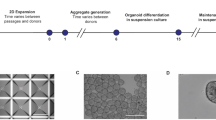
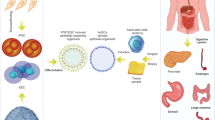
Abstract
Human epithelial organoids—3D spheroids derived from adult tissue stem cells—enable investigation of epithelial physiology and disease and host interactions with microorganisms, viruses and bioactive molecules. One challenge in using organoids is the difficulty in accessing the apical, or luminal, surface of the epithelium, which is enclosed within the organoid interior. This protocol describes a method we previously developed to control human and mouse organoid polarity in suspension culture such that the apical surface faces outward to the medium (apical-out organoids). Our protocol establishes apical-out polarity rapidly (24–48 h), preserves epithelial integrity, maintains secretory and absorptive functions and allows regulation of differentiation. Here, we provide a detailed description of the organoid polarity reversal method, compatible characterization assays and an example of an application of the technology—specifically the impact of host–microbe interactions on epithelial function. Control of organoid polarity expands the possibilities of organoid use in gastrointestinal and respiratory health and disease research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and in our primary research article21. No datasets were generated or analyzed during the current study. Source data are provided with this paper. Additional source data underlying the figures are available from the corresponding author upon request.
References
Barker, N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15, 19–33 (2014).
Sato, T. & Clevers, H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340, 1190–1194 (2013).
Huch, M. & Koo, B. K. Modeling mouse and human development using organoid cultures. Development 142, 3113–3125 (2015).
Clevers, H. Modeling development and disease with organoids. Cell 165, 1586–1597 (2016).
Han, X. et al. Lactobacillus rhamnosus GG prevents epithelial barrier dysfunction induced by interferon-gamma and fecal supernatants from irritable bowel syndrome patients in human intestinal enteroids and colonoids. Gut Microbes 10, 59–76 (2019).
Yin, Y., Jonge, H. R., Wu, X. & Yin, Y. Enteroids for nutritional studies. Mol. Nutr. Food Res. 63, e1801143 (2019).
Zachos, N. C. et al. Human enteroids/colonoids and intestinal organoids functionally recapitulate normal intestinal physiology and pathophysiology. J. Biol. Chem. 291, 3759–3766 (2016).
Pearce, S. C. et al. Intestinal enteroids recapitulate the effects of short-chain fatty acids on the intestinal epithelium. PLoS One 15, e0230231 (2020).
Saxena, K. et al. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J. Virol. 90, 43–56 (2016).
Zang, R. et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 5, eabc3582 (2020).
Koestler, B. J. et al. Human intestinal enteroids as a model system of Shigella pathogenesis. Infect. Immun. 87, e00733-18 (2019).
Sierra, J. C. et al. Spermine oxidase mediates Helicobacter pylori-induced gastric inflammation, DNA damage, and carcinogenic signaling. Oncogene 39, 4465–4474 (2020).
Ranganathan, S. et al. Evaluating Shigella flexneri pathogenesis in the human enteroid model. Infect. Immun. 87, e00740-18 (2019).
Bartfeld, S. et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148, 126–136 (2015).
Yoo, J.-H., Donowitz, M. & Yoo, J. H. Intestinal enteroids/organoids: a novel platform for drug discovery in inflammatory bowel diseases. World J. Gastroenterol. 25, 4125–4147 (2019).
Schütte, M. et al. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat. Commun. 8, 14262 (2017).
Van De Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Bigorgne, A. E. et al. TTC7A mutations disrupt intestinal epithelial apicobasal polarity. J. Clin. Invest. 124, 328–337 (2014).
Sato, T. et al. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011).
Co, J. Y. et al. Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep. 26, 2509–2520 (2019).
Fujii, M. & Sato, T. Somatic cell-derived organoids as prototypes of human epithelial tissues and diseases. Nat. Mater. 20, 156–169 (2021).
Salahudeen, A. A. et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 588, 670–675 (2020).
Krüger, M. et al. Cellulose nanofibril hydrogel promotes hepatic differentiation of human liver organoids. Adv. Healthc. Mater. 9, e1901658 (2020).
Li, Y. et al. Next-generation porcine intestinal organoids: an apical-out organoid model for swine enteric virus infection and immune response investigations. J. Virol. 94, e0100620 (2020).
Wilson, S. S., Tocchi, A., Holly, M. K., Parks, W. C. & Smith, J. G. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol. 8, 352–361 (2014).
Bartfeld, S. & Clevers, H. Organoids as model for infectious diseases: culture of human and murine stomach organoids and microinjection of Helicobacter pylori. J. Vis. Exp. 105, 53359 (2015).
Buti, L. et al. CagA–ASPP2 complex mediates loss of cell polarity and favors H. Pylori colonization of human gastric organoids. Proc. Natl Acad. Sci. USA 117, 2645–2655 (2020).
Jattan, J. et al. Using primary murine intestinal enteroids to study dietary TAG absorption, lipoprotein synthesis, and the role of apoC-III in the intestine. J. Lipid Res. 58, 853–865 (2017).
Wilson, S. S. et al. Alpha-defensin-dependent enhancement of enteric viral infection. PLoS Pathog. 13, e1006446 (2017).
Lamers, M. M. et al. SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54 (2020).
VanDussen, K. L. et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 64, 911–920 (2015).
Moon, C., Vandussen, K. L., Miyoshi, H. & Stappenbeck, T. S. Development of a primary mouse intestinal epithelial cell monolayer culture system to evaluate factors that modulate IgA transcytosis. Mucosal Immunol. 7, 818–828 (2014).
Williamson, I. A. et al. A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cell. Mol. Gastroenterol. Hepatol. 6, 301–319 (2018).
Fasciano, A. C., Blutt, S. E., Estes, M. K. & Mecsas, J. Induced differentiation of m cell-like cells in human stem cell-derived ileal enteroid monolayers. J. Vis. Exp. (149), e59894 (2019).
Ding, S. et al. Retinoic acid and lymphotoxin signaling promote differentiation of human intestinal M cells. Gastroenterology 159, 214–226.e1 (2020).
Kumar, P. et al. Enterotoxigenic Escherichia coli–blood group A interactions intensify diarrheal severity. J. Clin. Invest. 128, 3298–3311 (2018).
In, J. et al. Enterohemorrhagic Escherichia coli reduces mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell. Mol. Gastroenterol. Hepatol. 2, 48–62.e3 (2016).
Wosen, J. E. et al. Human intestinal enteroids model MHC-II in the gut epithelium. Front. Immunol. 10, 1970 (2019).
Pentecost, M., Kumaran, J., Ghosh, P. & Amieva, M. R. Listeria monocytogenes internalin B activates junctional endocytosis to accelerate intestinal invasion. PLoS Pathog. 6, e1000900 (2010).
Galán, J. E. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17, 53–86 (2001).
Rajan, A. et al. Enteroaggregative E. coli adherence to human heparan sulfate proteoglycans drives segment and host specific responses to infection. PLOS Pathog. 16, e1008851 (2020).
Giobbe, G. G. et al. SARS-CoV-2 infection and replication in human fetal and pediatric gastric organoids. Preprint at bioRxiv https://doi.org/10.1101/2020.06.24.167049 (2020).
Suzuki, T. Regulation of intestinal epithelial permeability by tight junctions. Cell. Mol. Life Sci. 70, 631–659 (2013).
Oshima, T. & Miwa, H. Gastrointestinal mucosal barrier function and diseases. J. Gastroenterol. 51, 768–778 (2016).
Marchiando, A. M., Graham, W. V. & Turner, J. R. Epithelial barriers in homeostasis and disease. Annu. Rev. Pathol. 5, 119–144 (2010).
Pagano, R. E. & Sleight, R. G. Defining lipid transport pathways in animal cells. Science 229, 1051–1057 (1985).
Wang, T. Y., Liu, M., Portincasa, P. & Wang, D. Q. H. New insights into the molecular mechanism of intestinal fatty acid absorption. Eur. J. Clin. Invest. 43, 1203–1223 (2013).
Dubikovskaya, E., Chudnovskiy, R., Karateev, G., Park, H. M. & Stahl, A. Measurement of long-chain fatty acid uptake into adipocytes. Methods Enzymol. 538, 107–134 (2014).
Drecktrah, D. et al. Dynamic behavior of Salmonella-induced membrane tubules in epithelial cells. Traffic 9, 2117–2129 (2008).
Miyoshi, H. & Stappenbeck, T. S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 8, 2471–2482 (2013).
Acknowledgements
We thank R. Cooper for reagent preparation and the laboratory of C. Kuo for support in deriving GI organoids. This study was supported by the Stanford Child Health Research Institute Postdoctoral Award (to J.Y.C.), NIH T32AI007328-29 (support to J.Y.C.), Novo Nordisk Foundation Challenge Programme (to M.R.A and M.M.-C.), NIH U19AI116484-01 (to M.R.A. and D.M.M.), Bill and Melinda Gates Foundation/Stanford CHSI Pilot Grant (to M.R.A. and D.M.M) and OPP1113682 (to M.R.A).
Author information
Authors and Affiliations
Contributions
J.Y.C. and M.M.-C developed the protocol for the apical-out GI organoids and the referenced apical-out lung organoids, respectively, and performed the experiments. J.Y.C., M.M.-C., D.M.M. and M.R.A. conceived the study and prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Co, J. Y. et al. Cell Rep. 26, 2509–2520 (2019): https://doi.org/10.1016/j.celrep.2019.01.108
Salahudeen, A. A. et al. Nature 588, 670–675 (2020): https://doi.org/10.1038/s41586-020-3014-1
Supplementary information
41596_2021_607_MOESM1_ESM.mov
Supplementary Video 1 Human gastroid polarity reversal. Time-lapse DIC microscopy movie of a gastroid immediately after removal from BME shows that the process of polarity reversal occurs by morphogenetic eversion. The same gastroid was fixed, stained and imaged by 3D confocal microscopy to confirm apical-out polarity (F-actin–rich microvilli (arrow) face outward). Nuclei are stained with DAPI (blue; ThermoFisher Scientific, cat. no. D3571; RRID: AB_2307445), and the actin cytoskeleton is stained with AlexaFluor 660 phalloidin (F-actin, white; ThermoFisher, cat. no. A22285), both diluted 1:500. Scale bar, 10 µm.
Source data
Source Data Fig. 3
Raw data Fig. 3.
Source Data Fig. 4
Raw data Fig. 4b.
Source Data Fig. 5
Raw data Fig. 5c.
Rights and permissions
About this article
Cite this article
Co, J.Y., Margalef-Català, M., Monack, D.M. et al. Controlling the polarity of human gastrointestinal organoids to investigate epithelial biology and infectious diseases. Nat Protoc 16, 5171–5192 (2021). https://doi.org/10.1038/s41596-021-00607-0
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41596-021-00607-0
This article is cited by
-
Biomimetic culture substrates for modelling homeostatic intestinal epithelium in vitro
Nature Communications (2025)
-
Lactoferrin-osteopontin complexes: insights into intestinal organoid bioavailability and gut microbiota modulation
Communications Biology (2025)
-
Model systems to study tumor-microbiome interactions in early-onset colorectal cancer
EMBO Molecular Medicine (2025)
-
A scalable gut epithelial organoid model reveals the genome-wide colonization landscape of a human-adapted pathogen
Nature Genetics (2025)
-
Visualizing the internalization and biological impact of nanoplastics in live intestinal organoids by Fluorescence Lifetime Imaging Microscopy (FLIM)
Light: Science & Applications (2025)