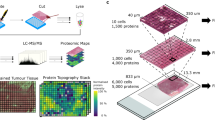
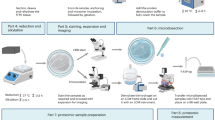
Abstract
High-throughput lysis and proteolytic digestion of biopsy-level tissue specimens is a major bottleneck for clinical proteomics. Here we describe a detailed protocol of pressure cycling technology (PCT)-assisted sample preparation for proteomic analysis of biopsy tissues. A piece of fresh frozen or formalin-fixed paraffin-embedded tissue weighing ~0.1–2 mg is placed in a 150 μL pressure-resistant tube called a PCT-MicroTube with proper lysis buffer. After closing with a PCT-MicroPestle, a batch of 16 PCT-MicroTubes are placed in a Barocycler, which imposes oscillating pressure to the samples from one atmosphere to up to ~3,000 times atmospheric pressure. The pressure cycling schemes are optimized for tissue lysis and protein digestion, and can be programmed in the Barocycler to allow reproducible, robust and efficient protein extraction and proteolysis digestion for mass spectrometry-based proteomics. This method allows effective preparation of not only fresh frozen and formalin-fixed paraffin-embedded tissue, but also cells, feces and tear strips. It takes ~3 h to process 16 samples in one batch. The resulting peptides can be analyzed by various mass spectrometry-based proteomics methods. We demonstrate the applications of this protocol with mouse kidney tissue and eight types of human tumors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The raw MS data of this study have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository40 with the dataset identifier PXD030645. Project accession of the mouse kidney data: IPX0003852001. Project accession of the cancer tissue data: IPX0003852002.
References
Aebersold, R. & Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016).
Zhu, Y., Aebersold, R., Mann, M. & Guo, T. SnapShot: clinical proteomics. Cell 184, 4840–4840.e1 (2021).
Xiao, Q. et al. High-throughput proteomics and AI for cancer biomarker discovery. Adv. Drug Deliv. Rev. 176, 113844 (2021).
Guo, T. et al. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nat. Med. 21, 407–413 (2015).
Zhu, Y. et al. Identification of protein abundance changes in hepatocellular carcinoma tissues using PCT-SWATH. Proteomics Clin. Appl. 13, 1700179 (2019).
Eckert, M. A. et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature 569, 723–728 (2019).
Hood, B. L. et al. Proteomic analysis of formalin-fixed prostate cancer tissue. Mol. Cell Proteomics 4, 1741–1753 (2005).
Nie, X. et al. Multi-organ proteomic landscape of COVID-19 autopsies. Cell 184, 775–791.e14 (2021).
Wisniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
Hwang, S. I. et al. Direct cancer tissue proteomics: a method to identify candidate cancer biomarkers from formalin-fixed paraffin-embedded archival tissues. Oncogene 26, 65–76 (2007).
Hughes, C. S. et al. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 10, 757 (2014).
Hughes, C. S. et al. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 14, 68–85 (2019).
Fowler, C. B. et al. Elevated hydrostatic pressure promotes protein recovery from formalin-fixed, paraffin-embedded tissue surrogates. Lab Invest. 88, 185–195 (2008).
Powell, B. S., Lazarev, A. V., Carlson, G., Ivanov, A. R. & Rozak, D. A. Pressure cycling technology in systems biology. Methods Mol. Biol. 881, 27–62 (2012).
Shao, S. et al. Minimal sample requirement for highly multiplexed protein quantification in cell lines and tissues by PCT-SWATH mass spectrometry. Proteomics 15, 3711–3721 (2015).
Shao, S. et al. Reproducible tissue homogenization and protein extraction for quantitative proteomics using MicroPestle-assisted pressure-cycling technology. J. Proteome Res. 15, 1821–1829 (2016).
Thompson, A. et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 (2003).
Ross, P. L. et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics 3, 1154–1169 (2004).
Anderson, L. & Hunter, C. L. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell Proteomics 5, 573–588 (2006).
Peterson, A. C., Russell, J. D., Bailey, D. J., Westphall, M. S. & Coon, J. J. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell Proteomics 11, 1475–1488 (2012).
Gallien, S. et al. Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol. Cell Proteomics 11, 1709–1723 (2012).
Venable, J. D., Dong, M. Q., Wohlschlegel, J., Dillin, A. & Yates, J. R. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 (2004).
Powell, K. Technology to watch in 2018. Nature 553, 531–534 (2018).
Zhang, F., Ge, W., Ruan, G., Cai, X. & Guo, T. Data-independent acquisition mass spectrometry-based proteomics and software tools: a glimpse in 2020. Proteomics 20, 1900276 (2021).
Gao, H. et al. Accelerated lysis and proteolytic digestion of biopsy-level fresh-frozen and FFPE tissue samples using pressure cycling technology. J. Proteome Res. 19, 1982–1990 (2020).
Zhu, Y. & Guo, T. High-throughput proteomic analysis of fresh-frozen biopsy tissue samples using pressure cycling technology coupled with SWATH Mass spectrometry. Methods Mol. Biol. 1788, 279–287 (2018).
Zhu, Y. et al. High-throughput proteomic analysis of FFPE tissue samples facilitates tumor stratification. Mol. Oncol. 13, 2305–2328 (2019).
Zhu, T. et al. DPHL: a DIA pan-human protein mass spectrometry library for robust biomarker discovery. Genomics Proteomics Bioinformatics 18, 104–119 (2020).
Ge, W. et al. Computational optimization of spectral library size improves DIA-MS proteome coverage and applications to 15 tumors. J. Proteome Res. 20, 5392–5401 (2021).
Cheng, Y., Chen, Y. & Yu, C. Fast and efficient non-reduced Lys-C digest using pressure cycling technology for antibody disulfide mapping by LC–MS. J. Pharm. Biomed. Anal. 129, 203–209 (2016).
Nie, S., Greer, T., Huang, X., Zheng, X. & Li, N. Development of a simple non-reduced peptide mapping method that prevents disulfide scrambling of mAbs without affecting tryptic enzyme activity. J. Pharm. Biomed. Anal. 209, 114541 (2022).
Huang, Y., Burchmore, R., Jonsson, N. N., Johnson, P. C. D. & Eckersall, P. D. Technical report: In-gel sample preparation prior to proteomic analysis of bovine faeces increases protein identifications by removal of high molecular weight glycoproteins. J. Proteomics 261, 104573 (2022).
Wu, C. et al. Coupling suspension trapping-based sample preparation and data-independent acquisition mass spectrometry for sensitive exosomal proteomic analysis. Anal. Bioanal. Chem. 414, 2585–2595 (2022).
Xuan, Y. et al. Standardization and harmonization of distributed multi-center proteotype analysis supporting precision medicine studies. Nat. Commun. 11, 5248 (2020).
Hunt, A. L. et al. Extensive three-dimensional intratumor proteomic heterogeneity revealed by multiregion sampling in high-grade serous ovarian tumor specimens. iScience 24, 102757 (2021).
Lee, S. et al. Molecular analysis of clinically defined subsets of high-grade serous ovarian cancer. Cell Rep. 31, 107502 (2020).
Li, D. et al. pFind: a novel database-searching software system for automated peptide and protein identification via tandem mass spectrometry. Bioinformatics 21, 3049–3050 (2005).
Demichev, V., Messner, C. B., Vernardis, S. I., Lilley, K. S. & Ralser, M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44 (2020).
Cai, X. et al. PulseDIA: data-independent acquisition mass spectrometry using multi-injection pulsed gas-phase fractionation. J. Proteome Res. 20, 279–288 (2021).
Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res. 47, D1211–D1217 (2019).
Acknowledgements
This work is supported by grants from the National Key R&D Program of China (2021YFA1301601 and 2020YFE0202200), Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholars (LR19C050001), National Natural Science Foundation of China (81972492) and National Science Fund for Young Scholars (21904107).
Author information
Authors and Affiliations
Contributions
T.G. and X.C. designed the studies. X.C., X.Y., W.L., C.C., H.G., J.Y. and L.X. participated in the development of this protocol. X.C., C.L., R.S, L.Q. and L.Y. performed the PCT-based sample preparation. X.C performed the LC–MS/MS analysis. X.C., Z.X. and W.G. analyzed the data. X.C., T.G. and Y.Z. wrote the manuscript with input from all co-authors. T.G. and Y.Z. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
Y.Z. and T.G. are shareholders of Westlake Omics Inc. C.W., W.G., X.Y., W.L., C.C., H.G., J.Y. and L.X. are employees of Westlake Omics Inc. The other authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Louise Bundgaard and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Guo, T. et al. Nat. Med. 21, 407–413 (2015): https://doi.org/10.1038/nm.3807
Zhu, Y. et al. Mol. Oncol. 13, 2305–2328 (2019): https://doi.org/10.1002/1878-0261.12570
Shao, W. et al. Nat. Commun. 10, 2524 (2019): https://doi.org/10.1038/s41467-019-10513-5
Charmpi, K. et al. Genome Biol. 21, 302 (2020): https://doi.org/10.1186/s13059-020-02188-9
Nie, X. et al. Cell 184, 775–791.e714 (2021): https://doi.org/10.1016/j.cell.2021.01.004
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Supplementary Data 1
Experimental data from Fig. 2. Proteomic data of mouse kidney samples in the form of FF, FFPE punch and FFPE slice.
Supplementary Data 2
Experimental data from Fig. 3. Proteomic data of 32 independent FFPE tissue samples (benign and tumor pairs) from eight types of cancer tissues.
Supplementary Data 3
PulseDIA-PASEF MS isolation window: part 1 setting.
Supplementary Data 4
PulseDIA-PASEF MS isolation window: part 2 setting.
Supplementary Video
The procedures for handling the PCT-MicroTubes, MicroCaps, MicroPestles with the PCT tool.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cai, X., Xue, Z., Wu, C. et al. High-throughput proteomic sample preparation using pressure cycling technology. Nat Protoc 17, 2307–2325 (2022). https://doi.org/10.1038/s41596-022-00727-1
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41596-022-00727-1
This article is cited by
-
Mass-spectrometry-based proteomics: from single cells to clinical applications
Nature (2025)
-
iDIA-QC: AI-empowered data-independent acquisition mass spectrometry-based quality control
Nature Communications (2025)
-
A protein-based classifier for differentiating follicular thyroid adenoma and carcinoma
EMBO Molecular Medicine (2025)
-
A single-sample workflow for joint metabolomic and proteomic analysis of clinical specimens
Clinical Proteomics (2024)
-
AI-driven eyelid tumor classification in ocular oncology using proteomic data
npj Precision Oncology (2024)