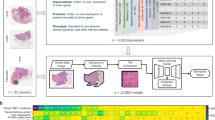
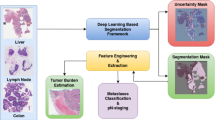
Abstract
Hematoxylin- and eosin-stained whole-slide images (WSIs) are the foundation of diagnosis of cancer. In recent years, development of deep learning-based methods in computational pathology has enabled the prediction of biomarkers directly from WSIs. However, accurately linking tissue phenotype to biomarkers at scale remains a crucial challenge for democratizing complex biomarkers in precision oncology. This protocol describes a practical workflow for solid tumor associative modeling in pathology (STAMP), enabling prediction of biomarkers directly from WSIs by using deep learning. The STAMP workflow is biomarker agnostic and allows for genetic and clinicopathologic tabular data to be included as an additional input, together with histopathology images. The protocol consists of five main stages that have been successfully applied to various research problems: formal problem definition, data preprocessing, modeling, evaluation and clinical translation. The STAMP workflow differentiates itself through its focus on serving as a collaborative framework that can be used by clinicians and engineers alike for setting up research projects in the field of computational pathology. As an example task, we applied STAMP to the prediction of microsatellite instability (MSI) status in colorectal cancer, showing accurate performance for the identification of tumors high in MSI. Moreover, we provide an open-source code base, which has been deployed at several hospitals across the globe to set up computational pathology workflows. The STAMP workflow requires one workday of hands-on computational execution and basic command line knowledge.
Key points
-
STAMP (solid tumor associative modeling in pathology) is a practical workflow for end-to-end weakly supervised deep learning in computational pathology, enabling prediction of biomarkers directly from whole-slide images.
-
This protocol differentiates itself from others by providing a collaborative framework through which clinical researchers can work with engineers to set up a complete computational pathology project.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Histopathology slides and genomics data from TCGA and CPTAC were used to train and validate the models. The slides for TCGA are available at https://portal.gdc.cancer.gov/. The slides for CPTAC are available at https://proteomics.cancer.gov/data-portal. The molecular and clinical data for TCGA and CPTAC used in the experiments are available at https://github.com/KatherLab/cancer-metadata. Source data are provided with this paper.
Code availability
The open-source STAMP software for the implementation of the MSI experiments is available on GitHub (https://github.com/KatherLab/STAMP).
References
Shmatko, A., Ghaffari Laleh, N., Gerstung, M. & Kather, J. N. Artificial intelligence in histopathology: enhancing cancer research and clinical oncology. Nat. Cancer 3, 1026–1038 (2022).
Ghaffari Laleh, N. et al. Benchmarking weakly-supervised deep learning pipelines for whole slide classification in computational pathology. Med. Image Anal. 79, 102474 (2022).
Foersch, S. et al. Deep learning for diagnosis and survival prediction in soft tissue sarcoma. Ann. Oncol. 32, 1178–1187 (2021).
Klein, C. et al. Artificial intelligence for solid tumour diagnosis in digital pathology. Br. J. Pharmacol. 178, 4291–4315 (2021).
Woerl, A.-C. et al. Deep learning predicts molecular subtype of muscle-invasive bladder cancer from conventional histopathological slides. Eur. Urol. 78, 256–264 (2020).
Hong, R., Liu, W., DeLair, D., Razavian, N. & Fenyö, D. Predicting endometrial cancer subtypes and molecular features from histopathology images using multi-resolution deep learning models. Cell Rep. Med. 2, 100400 (2021).
Kather, J. N. et al. Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. PLoS Med. 16, e1002730 (2019).
Ghaffari Laleh, N. et al. Deep Learning for interpretable end-to-end survival (E-ESurv) prediction in gastrointestinal cancer histopathology. Proceedings of the MICCAI Workshop on Computational Pathology. PMLR 156, 81–93 (2021).
Foersch, S. et al. Multistain deep learning for prediction of prognosis and therapy response in colorectal cancer. Nat. Med. 29, 430–439 (2023).
Wang, C.-W. et al. Weakly supervised deep learning for prediction of treatment effectiveness on ovarian cancer from histopathology images. Comput. Med. Imaging Graph. 99, 102093 (2022).
Ghaffari Laleh, N., Ligero, M., Perez-Lopez, R. & Kather, J. N. Facts and hopes on the use of artificial intelligence for predictive immunotherapy biomarkers in cancer. Clin. Cancer Res. 29, 316–323 (2023).
Kather, J. N. et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Cancer 1, 789–799 (2020).
Kanavati, F. et al. Weakly-supervised learning for lung carcinoma classification using deep learning. Sci. Rep. 10, 9297 (2020).
Wang, X. et al. Weakly supervised deep learning for whole slide lung cancer image analysis. IEEE Trans. Cybern. 50, 3950–3962 (2020).
Kather, J. N. et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 25, 1054–1056 (2019).
Bilal, M. et al. Development and validation of a weakly supervised deep learning framework to predict the status of molecular pathways and key mutations in colorectal cancer from routine histology images: a retrospective study. Lancet Digit. Health 3, e763–e772 (2021).
Schrammen, P. L. et al. Weakly supervised annotation-free cancer detection and prediction of genotype in routine histopathology. J. Pathol. 256, 50–60 (2022).
Echle, A. et al. Clinical-grade detection of microsatellite instability in colorectal tumors by deep learning. Gastroenterology 159, 1406–1416.e11 (2020).
Zeng, Q. et al. Artificial intelligence predicts immune and inflammatory gene signatures directly from hepatocellular carcinoma histology. J. Hepatol. 77, 116–127 (2022).
Jaroensri, R. et al. Deep learning models for histologic grading of breast cancer and association with disease prognosis. NPJ Breast Cancer 8, 113 (2022).
Li, C. et al. Weakly supervised mitosis detection in breast histopathology images using concentric loss. Med. Image Anal. 53, 165–178 (2019).
Zheng, Q. et al. A weakly supervised deep learning model and human-machine fusion for accurate grading of renal cell carcinoma from histopathology slides. Cancers (Basel) 15, 3198 (2023).
Muti, H. S. et al. The Aachen Protocol for Deep Learning Histopathology: A Hands-on Guide for Data Preprocessing. Available at https://oa.mg/work/10.5281/zenodo.3694994 (2020).
Graziani, M. et al. Attention-based interpretable regression of gene expression in histology. Interpretability of Machine Intelligence in Medical Image Computing: 5th International Workshop, iMIMIC 2022, Held in Conjunction with MICCAI 2022, Singapore, Singapore, September 22, 2022, Proceedings 44–60 (Springer-Verlag, 2022).
Campanella, G. et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25, 1301–1309 (2019).
Schmauch, B. et al. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat. Commun. 11, 1–15 (2020).
Lu, M. Y. et al. AI-based pathology predicts origins for cancers of unknown primary. Nature 594, 106–110 (2021).
Wagner, S. J. et al. Built to last? Reproducibility and reusability of deep learning algorithms in computational pathology. Mod. Pathol. 37, 100350 (2023).
Veldhuizen, G. P. et al. Deep learning-based subtyping of gastric cancer histology predicts clinical outcome: a multi-institutional retrospective study. Gastric Cancer 26, 708–720 (2023).
Muti, H. S. et al. Deep learning trained on lymph node status predicts outcome from gastric cancer histopathology: a retrospective multicentric study. Eur. J. Cancer 194, 113335 (2023).
Saldanha, O. L. et al. Direct prediction of genetic aberrations from pathology images in gastric cancer with swarm learning. Gastric Cancer 26, 264–274 (2023).
Niehues, J. M. et al. Generalizable biomarker prediction from cancer pathology slides with self-supervised deep learning: a retrospective multi-centric study. Cell Rep. Med. 4, 100980 (2023).
Wagner, S. J. et al. Transformer-based biomarker prediction from colorectal cancer histology: a large-scale multicentric study. Cancer Cell 41, 1650–1661.e4 (2023).
Jiang, X. et al. End-to-end prognostication in colorectal cancer by deep learning: a retrospective, multicentre study. Lancet Digit. Health 6, e33–e43 (2024).
Chatterji, S. et al. Prediction models for hormone receptor status in female breast cancer do not extend to males: further evidence of sex-based disparity in breast cancer. NPJ Breast Cancer 9, 91 (2023).
Hewitt, K. J. et al. Direct image to subtype prediction for brain tumors using deep learning. Neurooncol. Adv. 5, vdad139 (2023).
Saldanha, O. L. et al. Self-supervised attention-based deep learning for pan-cancer mutation prediction from histopathology. NPJ Precis. Oncol. 7, 35 (2023).
Loeffler, C. M. L. et al. Direct prediction of Homologous Recombination Deficiency from routine histology in ten different tumor types with attention-based Multiple Instance Learning: a development and validation study. Preprint at https://www.medrxiv.org/content/10.1101/2023.03.08.23286975v1 (2023).
El Nahhas, O. S. M. et al. Regression-based Deep-Learning predicts molecular biomarkers from pathology slides. Nat. Commun. 15, 1–253 (2024).
Wang, X. et al. Transformer-based unsupervised contrastive learning for histopathological image classification. Med. Image Anal. 81, 102559 (2022).
Causality in digital medicine. Nat. Commun. 12, 5471 (2021).
Wölflein, G. et al. Benchmarking pathology feature extractors for whole slide image classification. Preprint at https://arxiv.org/abs/2311.11772 (2023).
Goode, A., Gilbert, B., Harkes, J., Jukic, D. & Satyanarayanan, M. OpenSlide: a vendor-neutral software foundation for digital pathology. J. Pathol. Inform. 4, 27 (2013).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Ghassemi, M., Oakden-Rayner, L. & Beam, A. L. The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digit. Health 3, e745–e750 (2021).
Paszke, A. et al. PyTorch: An imperative style, high-performance deep learning library. In Advances in Neural Information Processing Systems (eds Wallach, H. et al.) Vol. 32 (Curran Associates, Inc., 2019).
Jorge Cardoso, M. et al. MONAI: an open-source framework for deep learning in healthcare. Preprint at https://arxiv.org/abs/2211.02701 (2022).
Martinez, K. & Cupitt, J. VIPS - a highly tuned image processing software architecture. In IEEE International Conference on Image Processing 2005. Genova, Italy II–574 (IEEE, 2005).
Janowczyk, A., Zuo, R., Gilmore, H., Feldman, M. & Madabhushi, A. HistoQC: an open-source quality control tool for digital pathology slides. JCO Clin. Cancer Inform. 3, 1–7 (2019).
Pedersen, A. et al. FastPathology: an open-source platform for deep learning-based research and decision support in digital pathology. IEEE Access 9, 58216–58229 (2021).
Pocock, J. et al. TIAToolbox as an end-to-end library for advanced tissue image analytics. Commun. Med. (Lond.) 2, 120 (2022).
Lu, M. Y. et al. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 5, 555–570 (2021).
Verghese, G. et al. Computational pathology in cancer diagnosis, prognosis, and prediction—present day and prospects. J. Pathol. 260, 551–563 (2023).
Saillard, C. et al. Validation of MSIntuit as an AI-based pre-screening tool for MSI detection from colorectal cancer histology slides. Nat. Commun. 14, 6695 (2023).
Greenson, J. K. et al. Pathologic predictors of microsatellite instability in colorectal cancer. Am. J. Surg. Pathol. 33, 126–133 (2009).
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Ellis, M. J. et al. Connecting genomic alterations to cancer biology with proteomics: the NCI Clinical Proteomic Tumor Analysis Consortium. Cancer Discov. 3, 1108–1112 (2013).
Macenko, M. et al. A method for normalizing histology slides for quantitative analysis. In 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro 1107–1110 (IEEE, 2009).
Howard, F. M. et al. The impact of site-specific digital histology signatures on deep learning model accuracy and bias. Nat. Commun. 12, 4423 (2021).
Canny, J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 8, 679–698 (1986).
Comes, M. C. et al. A deep learning model based on whole slide images to predict disease-free survival in cutaneous melanoma patients. Sci. Rep. 12, 20366 (2022).
Jiang, S., Suriawinata, A. A. & Hassanpour, S. MHAttnSurv: multi-head attention for survival prediction using whole-slide pathology images. Comput. Biol. Med. 158, 106883 (2023).
Sounderajah, V. et al. Developing a reporting guideline for artificial intelligence-centred diagnostic test accuracy studies: the STARD-AI protocol. BMJ Open 11, e047709 (2021).
Collins, G. S. et al. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open 11, e048008 (2021).
Trautmann, K. et al. Chromosomal instability in microsatellite-unstable and stable colon cancer. Clin. Cancer Res. 12, 6379–6385 (2006).
Lin, E. I. et al. Mutational profiling of colorectal cancers with microsatellite instability. Oncotarget 6, 42334–42344 (2015).
Boland, C. R. & Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 138, 2073–2087.e3 (2010).
Battaglin, F., Naseem, M., Lenz, H.-J. & Salem, M. E. Microsatellite instability in colorectal cancer: overview of its clinical significance and novel perspectives. Clin. Adv. Hematol. Oncol. 16, 735–745 (2018).
Selvaraju, R. R. et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. In 2017 IEEE International Conference on Computer Vision (ICCV) 618–626 (IEEE, 2017).
Pataki, B. Á. et al. HunCRC: annotated pathological slides to enhance deep learning applications in colorectal cancer screening. Sci. Data 9, 370 (2022).
Cheng, J. et al. Computational analysis of pathological images enables a better diagnosis of TFE3 Xp11.2 translocation renal cell carcinoma. Nat. Commun. 11, 1778 (2020).
Acknowledgements
We thank the testers of the protocol, S. Sainath, O. L. Saldanha, L. Žigutytė, C. Kummer, G. Serna, K. Boehm and L. Shaktah, who executed the STAMP protocol on various systems at cancer centers around the world. O.S.M.E.N. is supported by the German Federal Ministry of Education and Research (BMBF) through grant 1IS23070, Software Campus 3.0 (TU Dresden), as part of the Software Campus project ’MIRACLE-AI’. J.N.K. is supported by the German Federal Ministry of Health (DEEP LIVER, ZMVI1-2520DAT111), the German Cancer Aid (DECADE, 70115166), the German Federal Ministry of Education and Research (PEARL, 01KD2104C; CAMINO, 01EO2101; SWAG, 01KD2215A; TRANSFORM LIVER, 031L0312A; TANGERINE, 01KT2302 through ERA-NET Transcan), the German Academic Exchange Service (SECAI, 57616814), the German Federal Joint Committee (TransplantKI, 01VSF21048), the European Union’s Horizon Europe and innovation programme (ODELIA, 101057091; GENIAL, 101096312) and the National Institute for Health and Care Research (NIHR, NIHR213331) Leeds Biomedical Research Centre. G.W. is supported by Lothian NHS. D.T. is supported by the German Federal Ministry of Education and Research (SWAG, 01KD2215A; TRANSFORM LIVER) and the European Union’s Horizon Europe and innovation programme (ODELIA, 101057091). S.F. is supported by the German Federal Ministry of Education and Research (SWAG, 01KD2215A), the German Cancer Aid (DECADE, 70115166) and the German Research Foundation (504101714). S.J.W. was supported by the Helmholtz Association under the joint research school ‘Munich School for Data Science – MUDS’ and the Add-on Fellowship of the Joachim Herz Foundation. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care. This work was funded by the European Union. Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union. Neither the European Union nor the granting authority can be held responsible for them.
Author information
Authors and Affiliations
Contributions
O.S.M.E.N. and J.N.K. designed the protocol. O.S.M.E.N., M.v.T., G.W. and T.L. developed the software and wrote technical documentation. O.S.M.E.N., M.v.T., G.W., T.L., M.L., M.U., S.J.W., F.K., S.F. and D.T. tested the software. O.S.M.E.N., J.N.K. and K.J.H. interpreted and analyzed the data. All authors wrote and reviewed the protocol and approved the final version for submission.
Corresponding author
Ethics declarations
Competing interests
O.S.M.E.N., F.K. and D.T. hold shares in StratifAI GmbH. J.N.K. declares consulting services for Owkin, France; DoMore Diagnostics, Norway; Panakeia, UK,; Scailyte, Switzerland; Mindpeak, Germany; and Histofy, UK; furthermore, he holds shares in StratifAI GmbH, Germany, and has received honoraria for lectures by AstraZeneca, Bayer, Eisai, MSD, BMS, Roche, Pfizer and Fresenius. D.T. received honoraria for lectures by Bayer and holds shares in StratifAI GmbH, Germany. S.F. has received honoraria from MSD and BMS.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Wagner, S. J. et al. Cancer Cell 41, 1650–1661.e4 (2023): https://doi.org/10.1016/j.ccell.2023.08.002
El Nahhas, O. S. M. et al. Nat. Commun. 15, 1253 (2024): https://doi.org/10.1038/s41467-024-45589-1
Jiang, X. et al. Lancet Digit. Health 6, e33–e43 (2024): https://doi.org/10.1016/S2589-7500(23)00208-X
Hewitt, K. J. et al. Neurooncol. Adv. 5, vdad139 (2023): https://doi.org/10.1093/noajnl/vdad139
Saldanha, O. L. et al. npj Precis. Onc. 7, 35 (2023): https://doi.org/10.1038/s41698-023-00365-0
Supplementary information
Supplementary Information
Supplementary Text 1, Table 1 and Fig. 1
Source data
Source data
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
El Nahhas, O.S.M., van Treeck, M., Wölflein, G. et al. From whole-slide image to biomarker prediction: end-to-end weakly supervised deep learning in computational pathology. Nat Protoc 20, 293–316 (2025). https://doi.org/10.1038/s41596-024-01047-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-024-01047-2
This article is cited by
-
Geometric multi-instance learning for weakly supervised gastric cancer segmentation
npj Digital Medicine (2026)
-
KI in der Diagnostik – aus Sicht der Pathologie
Die Pathologie (2026)
-
Artificial intelligence in digital pathology diagnosis and analysis: technologies, challenges, and future prospects
Military Medical Research (2026)
-
NHP2 and PRPF4 are hub genes associated with the prognosis of colorectal cancer
BMC Cancer (2025)
-
Deep learning-based histomorphological subtyping and risk stratification of small cell lung cancer from hematoxylin and eosin-stained whole slide images
Genome Medicine (2025)