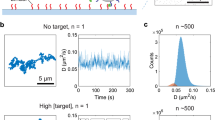
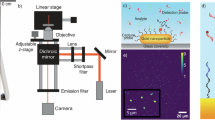
Abstract
The ability to apply controlled forces to individual molecules or molecular complexes and observe their behaviors has led to many important discoveries in biology. Instruments capable of probing single-molecule forces typically cost >US$100,000, limiting the use of these techniques. The centrifuge force microscope (CFM) is a low-cost and easy-to-use instrument that enables high-throughput single-molecule studies. By combining the imaging capabilities of a microscope with the force application of a centrifuge, the CFM enables the simultaneous probing of hundreds to thousands of single-molecule interactions using tethered particles. Here we present a comprehensive set of instructions for building a CFM module that fits within a commercial benchtop centrifuge. The CFM module uses a 3D-printed housing, relies on off-the-shelf optical and electrical components, and can be built for less than US$1,000 in about 1 day. We also provide detailed instructions for setting up and running an experiment to measure force-dependent shearing of a short DNA duplex, as well as the software for CFM control and data analysis. The protocol is suitable for users with basic experience in analytical biochemistry and biophysics. The protocol enables the use of CFM-based experiments and may facilitate access to the single-molecule research field.
Key points
-
The protocol covers the build and setup of a centrifuge force microscope that uses 3D-printed components and can be accommodated within an existing centrifuge, and the experimental procedure to probe the force-dependent dissociation of a short DNA duplex in the low piconewton force range.
-
Alternative methods include optical and magnetic tweezers, atomic force microscopy and acoustic forces, fluid flow or DNA-based probes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The main data discussed in this protocol (including raw source data that appear in Figs. 2 and 5) are available in the supporting primary research paper (https://doi.org/10.1038/s41467-023-36373-8). As mentioned in the data availability statement of the primary research paper, the raw videos are too large to be publicly shared (~1 TB) but are available for research purposes from the corresponding author upon reasonable request.
Code availability
Analysis software is available in the supporting primary research paper (https://doi.org/10.1038/s41467-023-36373-8). Our image acquisition software in LabView is provided in Supplementary Files.
References
Deniz, A. A., Mukhopadhyay, S. & Lemke, E. A. Single-molecule biophysics: at the interface of biology, physics and chemistry. J. R. Soc. Interface 5, 15–45 (2008).
Miller, H. et al. Single-molecule techniques in biophysics: a review of the progress in methods and applications. Rep. Prog. Phys. 81, 024601 (2017).
Sweeney, H. L. & Houdusse, A. Structural and functional insights into the myosin motor mechanism. Annu. Rev. Biophys. 39, 539–557 (2010).
Mohapatra, S. et al. Single-molecule analysis and engineering of DNA motors. Chem. Rev. 120, 36–78 (2019).
Seidel, R. & Dekker, C. Single-molecule studies of nucleic acid motors. Curr. Opin. Struct. Biol. 17, 80–86 (2007).
Woodside, M. T. & Block, S. M. Reconstructing folding energy landscapes by single-molecule force spectroscopy. Annu. Rev. Biophys. 43, 19–39 (2014).
Petrosyan, R., Narayan, A. & Woodside, M. T. Single-molecule force spectroscopy of protein folding. J. Mol. Biol. 433, 167207 (2021).
Mukherjee, S. et al. Single-molecule optical tweezers as a tool for delineating the mechanisms of protein-processing mechanoenzymes. ACS Omega 8, 87–97 (2022).
Camunas-Soler, J., Ribezzi-Crivellari, M. & Ritort, F. Elastic properties of nucleic acids by single-molecule force spectroscopy. Annu. Rev. Biophys. 45, 65–84 (2016).
Yodh, J. G., Schlierf, M. & Ha, T. Insight into helicase mechanism and function revealed through single-molecule approaches. Q. Rev. Biophys. 43, 185–217 (2010).
Smith, D. E. Single-molecule studies of viral DNA packaging. Curr. Opin. Virol. 1, 134–141 (2011).
Dulin, D. et al. Studying genomic processes at the single-molecule level: introducing the tools and applications. Nat. Rev. Genet. 14, 9–22 (2013).
Evans, E. Probing the relation between force—lifetime—and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 30, 105–128 (2001).
Puchner, E. M. & Gaub, H. E. Single-molecule mechanoenzymatics. Annu. Rev. Biophys. 41, 497–518 (2012).
Yusko, E. C. & Asbury, C. L. Force is a signal that cells cannot ignore. Mol. Biol. Cell 25, 3717–3725 (2014).
Eckels, E. C. et al. The work of titin protein folding as a major driver in muscle contraction. Annu. Rev. Physiol. 80, 327–351 (2018).
Neuman, K. C. & Nagy, A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 5, 491–505 (2008).
Moffitt, J. R. et al. Recent advances in optical tweezers. Annu. Rev. Biochem. 77, 205–228 (2008).
Polimeno, P. et al. Optical tweezers and their applications. J. Quant. Spectrosc. Radiat. Transf. 218, 131––150 (2018).
Bustamante, C. J. et al. Optical tweezers in single-molecule biophysics. Nat. Rev. Methods Prim. 1, 25 (2021).
De Vlaminck, I. & Dekker, C. Recent advances in magnetic tweezers. Annu. Rev. Biophys. 41, 453–472 (2012).
Kilinc, D. & Lee, G. U. Advances in magnetic tweezers for single molecule and cell biophysics. Integr. Biol. 6, 27–34 (2014).
Choi, H.-K. et al. High-resolution single-molecule magnetic tweezers. Annu. Rev. Biochem. 91, 33–59 (2022).
Engel, A. & Müller, D. J. Observing single biomolecules at work with the atomic force microscope. Nat. Struct. Biol. 7, 715–718 (2000).
Hinterdorfer, P. & Dufrêne, Y. F. Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods 3, 347–355 (2006).
Parot, P. et al. Past, present and future of atomic force microscopy in life sciences and medicine. J. Mol. Recogniti. 20, 418–431 (2007).
Ma, L. & Cockroft, S. L. Biological nanopores for single‐molecule biophysics. ChemBioChem 11, 25–34 (2010).
Sitters, G. et al. Acoustic force spectroscopy. Nat. Methods 12, 47–50 (2015).
Gourier, C. et al. A nanospring named erythrocyte. The biomembrane force probe. Cell. Mol. Bioeng. 1, 263–275 (2008).
Nickels, P. C. et al. Molecular force spectroscopy with a DNA origami-based nanoscopic force clamp. Science 354, 305–307 (2016).
Kim, S. et al. Multiplexed single-molecule assay for enzymatic activity on flow-stretched DNA. Nat. Methods 4, 397–399 (2007).
Akbari, E. et al. Low cost and massively parallel force spectroscopy with fluid loading on a chip. Nat. Commun. 13, 6800 (2022).
Halvorsen, K. & Wong, W. P. Massively parallel single-molecule manipulation using centrifugal force. Biophys. J. 98, L53––L55 (2010).
Yang, D. et al. Multiplexed single-molecule force spectroscopy using a centrifuge. Nat. Commun. 7, 11026 (2016).
Hoang, T., Patel, D. S. & Halvorsen, K. A wireless centrifuge force microscope (CFM) enables multiplexed single-molecule experiments in a commercial centrifuge. Rev. Sci. Instrum. 87, 083705 (2016).
Abraham Punnoose, J. et al. Wi-Fi live-streaming centrifuge force microscope for benchtop single-molecule experiments. Biophys. J. 119, 2231–2239 (2020).
Abraham Punnoose, J. et al. High-throughput single-molecule quantification of individual base stacking energies in nucleic acids. Nat. Commun. 14, 631 (2023).
Luo, Y. et al. Resolving molecular heterogeneity with single-molecule centrifugation. J. Am. Chem. Soc. 145, 3276–3282 (2023).
Kirkness, M. W. H. & Forde, N. R. Single-molecule assay for proteolytic susceptibility: force-induced collagen destabilization. Biophys. J. 114, 570–576 (2018).
LeFevre, T. B. et al. Measuring colloid–surface interaction forces in parallel using fluorescence centrifuge force microscopy. Soft Matter 17, 6326––6336 (2021).
Kou, L. et al. Real‐time parallel 3D multiple particle tracking with single molecule centrifugal force microscopy. J. Microsc. 273, 178–188 (2019).
Heinrich, V. et al. Imaging biomolecular interactions by fast three-dimensional tracking of laser-confined carrier particles. Langmuir 24, 1194–1203 (2008).
Rieu, M. et al. Parallel, linear, and subnanometric 3D tracking of microparticles with stereo darkfield interferometry. Sci. Adv. 7, eabe3902 (2021).
De Vlaminck, I. et al. Highly parallel magnetic tweezers by targeted DNA tethering. Nano Lett. 11, 5489–5493 (2011).
De Vlaminck, I. et al. Magnetic forces and DNA mechanics in multiplexed magnetic tweezers. PLoS One 7, e41432 (2012).
Ribeck, N. & Saleh, O. A. Multiplexed single-molecule measurements with magnetic tweezers. Rev. Sci. Instrum. 79, 094301 (2008).
Agarwal, R. & Duderstadt, K. E. Multiplex flow magnetic tweezers reveal rare enzymatic events with single molecule precision. Nat. Commun. 11, 4714 (2020).
Kamsma, D. & Wuite, G. J. L. Single-molecule measurements using acoustic force spectroscopy (AFS). In Single Molecule Analysis (ed. Peterman, E. et al.) 341–351 (Humana, 2018); https://doi.org/10.1007/978-1-4939-7271-5_18
Ott, W. et al. Single-molecule force spectroscopy on polyproteins and receptor–ligand complexes: the current toolbox. J. Struct. Biol. 197, 3–12 (2017).
Halvorsen, K., Schaak, D. & Wong, W. P. Nanoengineering a single-molecule mechanical switch using DNA self-assembly. Nanotechnology 22, 494005 (2011).
Yang, D. & Wong, W. P. Repurposing a benchtop centrifuge for high-throughput single-molecule force spectroscopy. In Single Molecule Analysis (ed. Peterman, E. et al.) 353–366 (Humana, 2018); https://doi.org/10.1007/978-1-4939-7271-5_19
Hoang, T., Moskwa, N. & Halvorsen, K. A ‘smart’tube holder enables real-time sample monitoring in a standard lab centrifuge. PLoS ONE 13, e0195907 (2018).
Dudko, O. K. Decoding the mechanical fingerprints of biomolecules. Q. Rev. Biophys. 49, e3 (2016).
Acknowledgements
The authors thank A. R. Chandrasekaran for useful conversations about the manuscript and figures, and B. Halvorsen for feedback to improve the CFM build instructions. Research reported in this publication was supported by the National Institutes of Health (NIH) through the National Institute of General Medical Sciences under awards R35GM124720 to K.H. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author information
Authors and Affiliations
Contributions
This protocol was conceived and planned by K.H. All authors were involved in different aspects of protocol development, primarily J.A.P. for experiment design and data analysis, C.S.K. for experiment preparation and running, A.H. for electrical design and 3D modeling, and K.H. for mechanical design and assembly. The main text was written by J.A.P. and K.H. with input and editing from all authors.
Corresponding author
Ethics declarations
Competing interests
K.H. has several issued patents and pending patent applications related to the CFM and has previously earned licensing royalties related to CFM.
Peer review
Peer review information
Nature Protocols thanks Hans Bergal, Wesley Wong and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key references
Abraham Punnoose, J. et al. High-throughput single-molecule quantification of individual base stacking energies in nucleic acids. Nat. Commun. 14.1, 631 (2023): https://doi.org/10.1038/s41467-023-36373-8
Abraham Punnoose, J. et al. Wi-Fi live-streaming centrifuge force microscope for benchtop single-molecule experiments. Biophys. J. 119.11, 2231–2239 (2020): https://doi.org/10.1016/j.bpj.2020.10.017
Supplementary information
Supplementary Information
Supplementary Notes 1–4, Figs. 1 and 2, Tables 1 and 2.
Supplementary Video 1
Video tutorial Steps 5–22.
Supplementary Video 2
Video tutorial Steps 23–38.
Supplementary Files
Fusion 3D model, individual .stl 3D models for printing, LabVIEW control software, MATLAB analysis code.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Abraham Punnoose, J., Hayden, A., Kam, C.S. et al. A guide to building a low-cost centrifuge force microscope module for single-molecule force experiments. Nat Protoc 20, 1951–1975 (2025). https://doi.org/10.1038/s41596-024-01102-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-024-01102-y