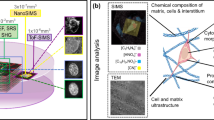
Abstract
Tissue microenvironments are extremely complex and heterogeneous. It is challenging to study metabolic interaction between the different cell types in a tissue with the techniques that are currently available. Here we describe a multimodal imaging pipeline that allows cell type identification and nanoscale tracing of stable isotope-labeled compounds. This pipeline extends upon the principles of correlative light, electron and ion microscopy, by combining confocal microscopy reporter or probe-based fluorescence, electron microscopy, stable isotope labeling and nanoscale secondary ion mass spectrometry. We apply this method to murine models of hepatocellular and mammary gland carcinomas to study uptake of glucose derived carbon (13C) and glutamine derived nitrogen (15N) by tumor-associated immune cells. In vivo labeling with fluorescent-tagged antibodies (B220, CD3, CD8a, CD68) in tandem with confocal microscopy allows for the identification of specific cell types (B cells, T cells and macrophages) in the tumor microenvironment. Subsequent image correlation with electron microscopy offers the contrast and resolution to image membranes and organelles. Nanoscale secondary ion mass spectrometry tracks the enrichment of stable isotopes within these intracellular compartments. The whole protocol described here would take ~6 weeks to perform from start to finish. Our pipeline caters to a broad spectrum of applications as it can easily be adapted to trace the uptake and utilization of any stable isotope-labeled nutrient, drug or a probe by defined cellular populations in any tissue in situ.
Key points
-
To understand complex tissues, it is useful to map metabolic processes to specific cell types. Nanoscale secondary ion mass spectrometry (NanoSIMS) can differentiate between isotopes derived from labeled metabolites at subcellular resolution.
-
This protocol combines three imaging modalities: confocal microscopy of fluorescent probes (antibodies), electron microscopy and NanoSIMS enabling cell type identification, characterization of the cellular architecture and extraction of metabolic information.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The raw images associated with the SEM and NanoSIMS for the different cell types (Fig. 2) are available at https://doi.org/10.25418/crick.24989841.
References
Kreuzaler, P. et al. Vitamin B5 supports MYC oncogenic metabolism and tumor progression in breast cancer. Nat. Metab. 5, 1870–1886 (2023).
Maclachlan, C., Sahlender, D. A., Hayashi, S., Molnár, Z. & Knott, G. Block face scanning electron microscopy of fluorescently labeled axons without using near infra-red branding. Front. Neuroanat. 12, 88 (2018).
Fearns, A., Greenwood, D. J., Rodgers, A., Jiang, H. & Gutierrez, M. G. Correlative light electron ion microscopy reveals in vivo localisation of bedaquiline in Mycobacterium tuberculosis-infected lungs. PLoS Biol. 18, e3000879 (2020).
de Boer, P., Hoogenboom, J. P. & Giepmans, B. N. G. Correlated light and electron microscopy: ultrastructure lights up! Nat. Methods 12, 503–513 (2015).
Arlauckas, S. P. et al. In vivo imaging reveals a tumor-associated macrophage–mediated resistance pathway in anti-PD-1 therapy. Sci. Transl. Med. 9, eaal3604 (2017).
Greenwood, D. J. et al. Subcellular antibiotic visualization reveals a dynamic drug reservoir in infected macrophages. Science 364, 1279–1282 (2019).
Méndez-Lucas, A. et al. Identifying strategies to target the metabolic flexibility of tumours. Nat. Metab. 2, 335–350 (2020).
Quinn, A. et al. Host-derived organic acids enable gut colonization of the honey bee symbiont Snodgrassella alvi. Nat. Microbiol. 9, 477–489 (2024).
Lechene, C. P., Luyten, Y., McMahon, G. & Distel, D. L. Quantitative imaging of nitrogen fixation by individual bacteria within animal cells. Science 317, 1563–1566 (2007).
Loussert-Fonta, C. et al. Correlation of fluorescence microscopy, electron microscopy, and NanoSIMS stable isotope imaging on a single tissue section. Commun. Biol. 3, 362 (2020).
Kreuzaler, P. et al. Heterogeneity of Myc expression in breast cancer exposes pharmacological vulnerabilities revealed through executable mechanistic modeling. Proc. Natl Acad. Sci. USA 116, 22399–22408 (2019).
Siuzdak, G. Subcellular quantitative imaging of metabolites at the organelle level. Nat. Metab. 5, 1446–1448 (2023).
Pfister, D. et al. NASH limits anti-tumour surveillance in immunotherapy-treated HCC. Nature 592, 450–456 (2021).
Kotsiliti, E. et al. Intestinal B cells license metabolic T-cell activation in NASH microbiota/antigen-independently and contribute to fibrosis by IgA-FcR signalling. J. Hepatol. 79, 296–313 (2023).
Garnelo, M. et al. Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut 66, 342–351 (2017).
Yuneva, M. O. et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 15, 157–170 (2012).
Tao, J. et al. Distinct anti-oncogenic effect of various microRNAs in different mouse models of liver cancer. Oncotarget 6, 6977–6988 (2015).
Lechene, C. et al. High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. J. Biol. 5, 20 (2006).
Titze, B., Genoud, C. & Friedrich, R. W. SBEMimage: versatile acquisition control software for serial block-face electron microscopy. Front. Neural Circuits 12, 54 (2018).
Bogovic, J. A., Hanslovsky, P., Wong, A. & Saalfeld, S. Robust registration of calcium images by learned contrast synthesis. In 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI) 1123–1126. https://doi.org/10.1109/ISBI.2016.7493463 (IEEE, 2016).
Valap for agar mounts. Cold Spring Harb Protoc. 2010, pdb.rec12380 (2010).
Deerinck, T. J., Bushong, E. A., Ellisman, M. H. & Thor, A. Preparation of biological tissues for serial block face scanning electron microscopy (SBEM). protocols.io https://doi.org/10.17504/protocols.io.36wgq7je5vk5/v2 (2022).
McMahon, G. & Lechene, C. P. High‐resolution multi‐isotope imaging mass spectrometry (MIMS) imaging applications in stem cell biology. Curr. Protoc. 1, e290 (2021).
Cardona, A. et al. TrakEM2 software for neural circuit reconstruction. PLoS ONE 7, e38011 (2012).
de Senneville, B. D. et al. Deciphering tumour tissue organization by 3D electron microscopy and machine learning. Commun. Biol. 4, 1390 (2021).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Argüello, R. J. et al. SCENITH: a flow cytometry-based method to functionally profile energy metabolism with single-cell resolution. Cell Metab. 32, 1063–1075.e7 (2020).
Hartmann, F. J. et al. Single-cell metabolic profiling of human cytotoxic T cells. Nat. Biotechnol. 39, 186–197 (2021).
Passarelli, M. K. et al. The 3D OrbiSIMS—label-free metabolic imaging with subcellular lateral resolution and high mass-resolving power. Nat. Methods 14, 1175–1183 (2017).
Kern, C. et al. Orbi‐SIMS mediated metabolomics analysis of pathogenic tissue up to cellular resolution. Chem. Methods https://doi.org/10.1002/cmtd.202400008 (2024).
Yan, X. et al. Cell-type-specific metabolic profiling achieved by combining desorption electrospray ionization mass spectrometry imaging and immunofluorescence staining. Anal. Chem. 92, 13281–13289 (2020).
Acknowledgements
This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK, the UK Medical Research Council and the Wellcome Trust FC001223 (M.Y.) and by the CRUK Grand Challenge Award 2015 C57633/A25043 (J.B. and M.Y.). We would like to thank all the animal technicians from the Francis Crick Institute’s Biological Research Facility for their dedicated work. LLMs (Microsoft Editor, ChatGPT and Grammarly) were solely used for grammar checks and reformatting purposes during the writing of this manuscript.
Author information
Authors and Affiliations
Contributions
S.V.V., P.K., C.M.L. and G.M. developed the protocol with critical input from M.Y., L.C. and J.B. The paper was written by S.V.V., P.K., C.M.L. and G.M. Further edits and suggestions for the manuscript were provided by G.G., L.C. and M.Y. The animal experiments and confocal microscopy imaging was performed by S.V.V. The sample preparation and EM imaging was carried out by C.M.L. All the NanoSIMS experiments and data analysis were performed by G.G. and G.M.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Anders Meibom and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key reference
Kreuzaler, P. et al. Nat. Metab. 5, 1870–1886 (2023): https://doi.org/10.1038/s42255-023-00915-7
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Venkateswaran, S.V., Kreuzaler, P., Maclachlan, C. et al. A multimodal imaging pipeline to decipher cell-specific metabolic functions and tissue microenvironment dynamics. Nat Protoc 20, 1678–1699 (2025). https://doi.org/10.1038/s41596-024-01118-4
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41596-024-01118-4