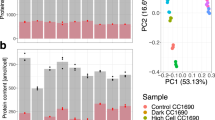
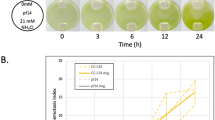
Abstract
The systematic cultivation of species of photosynthetically active ‘green’ microorganisms in research labs started in the 1940s. Among these microorganisms, Chlamydomonas represents a genus of green biciliated microalgae, of which Chlamydomonas reinhardtii has become the main describing species. For decades C. reinhardtii has been used as an established model organism in biology, including research areas such as molecular biology of eukaryotes, photosynthesis, light receptors, cell metabolism, the dynamics of microtubule assembly and protein transport along cilia. More recently, the use of suspensions of light-responsive living microorganisms has seen a major expansion from the life sciences to the biophysics, statistical physics, fluid dynamics and bioengineering communities. Studies that substantially advance the state of the art in these research areas require the reliable preparation and maintenance of viable, monodisperse and synchronous cell cultures. Although some technical aspects are shared with standard procedures in cell biology and microbiology, Chlamydomonas and its relatives are photosensitive and, simultaneously, motile, meaning this microorganism requires tailored cultivation protocols that are specific to this species. Here we provide guidance on which Chlamydomonas wild-type and mutant strains are suitable for specific experiments and provide detailed step-by-step procedures to measure culture synchronicity, growth rate of the population, average cell size and motility features. The reliable preparation of cell cultures may facilitate future interdisciplinary research using living suspensions of photoactive microorganisms.
Key points
-
Short-term Chlamydomonas cultures, prepared in liquid, are used for experiments, whereas long-term cultures are prepared on agar for the preservation of strains.
-
The preparation of synchronous suspensions of cells facilitates the reproducibility of data obtained in disciplines such as biophysics, statistical physics and fluid dynamics, where motility and collective behavior of large populations of cells is dependent on the health and synchronicity of the culture.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The source data supporting Figs. 4–8 can be retrieved via Zenodo at https://doi.org/10.5281/zenodo.11191785 (ref. 73).
Code availability
The code in MATLAB for cell detection, tracking and calculating the MSD is available via Zenodo at https://doi.org/10.5281/zenodo.13485141 (ref. 74) under Creative Commons Attribution v.4.0 International Public License and includes a user’s guide.
References
Pröschold, T., Marin, B., Schlösser, U. G. & Melkonian, M. Molecular phylogeny and taxonomic revision of Chlamydomonas (Chlorophyta). I. Emendation of Chlamydomonas Ehrenberg and Chloromonas Gobi, and description of Oogamochlamys gen. nov. and Lobochlamys gen. nov. Protist 152, 265–300 (2001).
Pröschold, T. & Darienko, T. in The Chlamydomonas Sourcebook Vol. 1 (ed. Goodenough, U.) Ch. 1 (Elsevier, 2023).
Gallaher, S. D., Fitz-Gibbon, S. T., Glaesener, A. G., Pellegrini, M. & Merchant, S. S. Chlamydomonas genome resource for laboratory strains reveals a mosaic of sequence variation, identifies true strain histories, and enables strain-specific studies. Plant Cell 27, 2335–2352 (2015).
Silflow, C. D. & Lefebvre, P. A. Assembly and motility of eukaryotic cilia and flagella. lessons from Chlamydomonas reinhardtii. Plant Physiol. 127, 1500–1507 (2001).
Rochaix, J.-D. Chlamydomonas reinhardtii as the photosynthetic yeast. Annu. Rev. Genet. 29, 209–230 (1995).
Hegemann, P. Algal sensory photoreceptors. Annu. Rev. Plant Biol. 59, 167–189 (2008).
Guschina, I. A. & Harwood, J. L. Lipids and lipid metabolism in eukaryotic algae. Prog. Lipid Res. 45, 160–186 (2006).
Silflow, C. D., Mackinder, L. C. & Wingfield, J. in The Chlamydomonas Sourcebook Vol. 1 (ed. Goodenough, U.) Ch. 14 (Elsevier, 2023).
Lorentzen, E. & Lechtreck, K. in The Chlamydomonas Sourcebook Vol. 3 (ed. Dutcher, S.) Ch. 12 (Elsevier, 2023).
Kreis, C. T., Le Blay, M., Linne, C., Makowski, M. M. & Bäumchen, O. Adhesion of Chlamydomonas microalgae to surfaces is switchable by light. Nat. Phys. 14, 45–49 (2018).
Laroussi, T., Jarrahi, M. & Amselem, G. Short-term memory effects in the phototactic behavior of microalgae. Soft Matter 20, 3996–4006 (2024).
Sjoblad, R. D. & Frederikse, P. H. Chemotactic responses of Chlamydomonas reinhardtii. Mol. Cell. Biol. 1, 1057–1060 (1981).
Fragkopoulos, A. A. et al. Self-generated oxygen gradients control collective aggregation of photosynthetic microbes. J. R. Soc. Interface 18, 20210553 (2021).
Goodenough, U. & Engel, B. D. in The Chlamydomonas Sourcebook Vol. 1 (ed. Goodenough, U.) Ch. 2 (Elsevier, 2023).
Schötz, F., Bathelt, H., Arnold, C.-G. & Schimmer, O. Die architektur und organisation der Chlamydomonas-zelle. Protoplasma 75, 229–254 (1972).
Boynton, J., Gillham, N. & Chabot, J. Chloroplast ribosome deficient mutants in the green alga Chlamydomonas reinhardi and the question of chloroplast ribosome function. J. Cell Sci. 10, 267–305 (1972).
Bloodgood, R. A. in The Chlamydomonas Sourcebook Vol. 3 (ed. Dutcher, S.) Ch. 10 (Elsevier, 2023).
Sale, W. S. & Dutcher, S. K. in The Chlamydomonas Sourcebook Vol. 3 (ed. Dutcher, S.) Ch. 1 (Elsevier, 2023).
Lechtreck, K. F. Ift–cargo interactions and protein transport in cilia. Trends Biochem. Sci. 40, 765–778 (2015).
Brown, J. M. & Witman, G. B. Cilia and diseases. BioScience 64, 1126–1137 (2014).
Umen, J. & Liu, D. in The Chlamydomonas Sourcebook Vol. 1 (ed. Goodenough, U.) Ch. 8 (Elsevier, 2023).
Coleman, A. W. The nuclear cell cycle in Chlamydomonas (Chlorophyceae) 1. J. Phycol. 18, 192–195 (1982).
Harper, J. & John, P. Coordination of division events in the Chlamydomonas cell cycle. Protoplasma 131, 118–130 (1986).
Craigie, R. & Cavalier-Smith, T. Cell volume and the control of the Chlamydomonas cell cycle. J. Cell Sci. 54, 173–191 (1982).
Goodenough, U. & Lee, J.-H. in The Chlamydomonas Sourcebook Vol. 1 Ch. 3 (ed. Goodenough, U.) 41–64 (Elsevier, 2023).
Schlösser, U. Enzymatisch gesteuerte Freisetzung von Zoosporen bei Chlamydomonas reinhardii Dangeard in Synchronkultur. Arch. Microbiol. 54, 129–159 (1966).
Goodenough, U., Lee, J.-H. & Snell, W. J. in The Chlamydomonas Sourcebook Vol. 1 (ed. Goodenough, U.) Ch. 9 (Elsevier, 2023).
Hui, C., Schmollinger, S. & Glaesener, A. G. in The Chlamydomonas Sourcebook Vol. 1 (ed. Goodenough, U.) Ch. 11 (Elsevier, 2023).
Sager, R. & Granick, S. Nutritional studies with Chlamydomonas reinhardi. Ann. N. Y. Acad. Sci. 56, 831–838 (1953).
Ma, F., Salomé, P. A., Merchant, S. S. & Pellegrini, M. Single-cell RNA sequencing of batch Chlamydomonas cultures reveals heterogeneity in their diurnal cycle phase. Plant Cell 33, 1042–1057 (2021).
Calatrava, V., Tejada-Jimenez, M., Sanz-Luque, E., Fernandez, E. & Galvan, A. in The Chlamydomonas Sourcebook Vol. 2 (eds. Grossman, A. R. & Wollman, F.-A.) Ch. 3 (Elsevier, 2023).
Pröschold, T., Harris, E. H. & Coleman, A. W. Portrait of a species: Chlamydomonas reinhardtii. Genetics 170, 1601–1610 (2005).
Gorman, D. S. & Levine, R. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl Acad. Sci. USA 54, 1665–1669 (1965).
Kropat, J. et al. A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 66, 770–780 (2011).
Terauchi, A. M., Peers, G., Kobayashi, M. C., Niyogi, K. K. & Merchant, S. S. Trophic status of Chlamydomonas reinhardtii influences the impact of iron deficiency on photosynthesis. Photosynth. Res. 105, 39–49 (2010).
Ostapenko, T. et al. Curvature-guided motility of microalgae in geometric confinement. Phys. Rev. Lett. 120, 068002 (2018).
Jin, D., Kotar, J., Silvester, E., Leptos, K. C. & Croze, O. A. Diurnal variations in the motility of populations of biflagellate microalgae. Biophys. J. 119, 2055–2062 (2020).
Cammann, J. et al. Emergent probability fluxes in confined microbial navigation. Proc. Natl Acad. Sci. USA 118, e2024752118 (2021).
Bruce, V. G. The biological clock in Chlamydomonas reinhardi. J. Protozool. 17, 328–334 (1970).
Arrieta, J., Barreira, A., Chioccioli, M., Polin, M. & Tuval, I. Phototaxis beyond turning: persistent accumulation and response acclimation of the microalga Chlamydomonas reinhardtii. Sci. Rep. 7, 3447 (2017).
Byrne, T. E., Wells, M. R. & Johnson, C. H. Circadian rhythms of chemotaxis to ammonium and of methylammonium uptake in Chlamydomonas. Plant Physiol. 98, 879–886 (1992).
Choi, H. I., Kim, J. Y. H., Kwak, H. S., Sung, Y. J. & Sim, S. J. Quantitative analysis of the chemotaxis of a green alga, Chlamydomonas reinhardtii, to bicarbonate using diffusion-based microfluidic device. Biomicrofluidics 10, 014121 (2016).
Straley, S. C. & Bruce, V. G. Stickiness to glass: circadian changes in the cell surface of Chlamydomonas reinhardi. Plant Physiol. 63, 1175–1181 (1979).
Kreis, C. T., Grangier, A. & Bäumchen, O. In vivo adhesion force measurements of Chlamydomonas on model substrates. Soft Matter 15, 3027–3035 (2019).
Till, S., Ebmeier, F., Fragkopoulos, A. A., Mazza, M. G. & Bäumchen, O. Motility and self-organization of gliding Chlamydomonas populations. Phys. Rev. Res. 4, L042046 (2022).
Cavalier-Smith, T. Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardii. J. Cell Sci. 16, 529–556 (1974).
Mittag, M., Kiaulehn, S. & Johnson, C. H. The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to? Plant Physiol. 137, 399–409 (2005).
Böddeker, T. J., Karpitschka, S., Kreis, C. T., Magdelaine, Q. & Bäumchen, O. Dynamic force measurements on swimming Chlamydomonas cells using micropipette force sensors. J. R. Soc. Interface 17, 20190580 (2020).
Kantsler, V., Dunkel, J., Polin, M. & Goldstein, R. E. Ciliary contact interactions dominate surface scattering of swimming eukaryotes. Proc. Natl Acad. Sci. USA 110, 1187–1192 (2013).
Williams, C. R. & Bees, M. A. A tale of three taxes: photo-gyro-gravitactic bioconvection. J. Exp. Biol. 214, 2398–2408 (2011).
Crutchfield, A., Diller, K. & Brand, J. Cryopreservation of Chlamydomonas reinhardtii (Chlorophyta). Eur. J. Phycol. 34, 43–52 (1999).
Day, J. G. Cryopreservation of microalgae and cyanobacteria. Methods Mol. Biol. 141–151 (2007).
Piasecki, B. P., Diller, K. R. & Brand, J. J. Cryopreservation of Chlamydomonas reinhardtii: a cause of low viability at high cell density. Cryobiology 58, 103–109 (2009).
Hlavová, M., Vítová, M. & Bišová, K. Synchronization of green algae by light and dark regimes for cell cycle and cell division studies. Methods Mol. Biol. 1370, 3–16 (2016).
Zhang, N. et al. Systems-wide analysis revealed shared and unique responses to moderate and acute high temperatures in the green alga Chlamydomonas reinhardtii. Commun. Biol. 5, 460 (2022).
Order Chlamydomonas strains from Chlamydomonas resource center (CC). Strains Archives https://www.chlamycollection.org/products/strains/ (2025).
Order Chlamydomonas strains from Sammlung von Algenkulturen Göttingen (SAG). MBM ScienceBridge https://sciencebridge.de/en/algae.html (2025).
Damoo, D. Y. & Durnford, D. G. Long-term survival of Chlamydomonas reinhardtii during conditional senescence. Arch. Microbiol. 203, 5333–5344 (2021).
Guide for cell counting using a hemocytometer (Sigma Aldrich). Milipore Sigma https://www.sigmaaldrich.com/DE/en/technical-documents/technical-article/cell-culture-and-cell-culture-analysis/mammalian-cell-culture/cell-quantification (2025).
Catalan, R. E. et al. Light-regulated adsorption and desorption of Chlamydomonas cells at surfaces. Soft Matter 19, 306–314 (2023).
Bassi, R., Soen, S. Y., Frank, G., Zuber, H. & Rochaix, J. Characterization of chlorophyll a/b proteins of photosystem I from Chlamydomonas reinhardtii. J. Biol. Chem. 267, 25714–25721 (1992).
Surzycki, S. in Methods in Enzymology Vol. 23 (ed. Pietro, A. S.) 67–73 (Elsevier, 1971).
Blair, D. & Dufresne, E. The matlab particle tracking code repository. Matlab http://physics.georgetown.edu/matlab (2008).
Kessler, J. O. Individual and collective fluid dynamics of swimming cells. J. Fluid Mech. 173, 191–205 (1986).
Cooper, M. B. & Smith, A. G. Exploring mutualistic interactions between microalgae and bacteria in the omics age. Curr. Opin. Plant Biol. 26, 147–153 (2015).
Aiyar, P. et al. Antagonistic bacteria disrupt calcium homeostasis and immobilize algal cells. Nat. Commun. 8, 1756 (2017).
Carrasco Flores, D. et al. A mutualistic bacterium rescues a green alga from an antagonist. Proc. Natl Acad. Sci. USA 121, e2401632121 (2024).
Kawachi, M. & Noël, M.-H. in Algal Culturing Techniques (ed. Andersen, R. A.) 65–81 (Elsevier, 2005).
Guillard, R. R. in Algal Culturing Techniques (ed. Andersen, R. A.) 117–132 (Elsevier, 2005).
Meeuse, B. A simple method for concentrating phototactic flagellates and separating them from debris. Arch. Mikrobiol. 45, 423–424 (1963).
Sieracki, M., Poulton, N. & Crosbie, N. in Algal Culturing Techniques (ed. Andersen, R. A.) 101–116 (Elsevier, 2005).
Kim, H. S., Devarenne, T. P. & Han, A. Microfluidic systems for microalgal biotechnology: a review. Algal Res. 30, 149–161 (2018).
Fragkopoulos, A. & Catalan, R. Cultivation of Chlamydomonas cells. Zenodo https://doi.org/10.5281/zenodo.11191785 (2024).
Fragkopoulos, A. & Catalan, R. Detection and tracking of Chlamydomonas cells. Zenodo https://doi.org/10.5281/zenodo.13485141 (2024).
Morishita, J., Tokutsu, R., Minagawa, J., Hisabori, T. & Wakabayashi, K.-I. Characterization of Chlamydomonas reinhardtii mutants that exhibit strong positive phototaxis. Plants 10, 1483 (2021).
Ide, T. et al. Identification of the agg1 mutation responsible for negative phototaxis in a “wild-type” strain of Chlamydomonas reinhardtii. Biochem. Biophys. Rep. 7, 379–385 (2016).
Claes, H. Autolyse der Zellwand bei den Gameten von Chlamydomonas reinhardii. Arch. Mikrobiol 78, 180–188 (1971).
Acknowledgements
The authors thank the Göttingen Algae Culture Collection (SAG) for providing SAG-labeled microalgal strains used in this work and T. Pröschold for insightful discussions. R.E.C. acknowledges generous financial support from the German Academic Exchange Service (DAAD).
Author information
Authors and Affiliations
Contributions
All authors contributed to the development of the protocol. R.E.C., A.A.F. and O.B. led the data analysis. R.E.C. and A.A.F. wrote the first draft of the manuscript. All authors contributed to the discussions and the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Tyler W. Johannes and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key References
Kreis, C. T. et al. Nat. Phys. 14, 45–49 (2018): https://doi.org/10.1038/nphys4258
Cammann, J. et al. Proc. Natl Acad. Sci. USA 118, e2024752118 (2021): https://doi.org/10.1073/pnas.2024752118
Ostapenko, T. et al. Phys. Rev. Lett. 120, 068002 (2018): https://doi.org/10.1103/PhysRevLett.120.068002
Böddeker, T. J. et al. J. R. Soc. Interface 17, 20190580 (2020): https://doi.org/10.1098/rsif.2019.0580
Till, S. et al. Phys. Rev. Res 4, L042046 (2022): https://doi.org/10.1103/PhysRevResearch.4.L042046
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Catalan, R.E., Fragkopoulos, A.A., Girot, A. et al. Preparation, maintenance and propagation of synchronous cultures of photoactive Chlamydomonas cells. Nat Protoc 20, 2125–2150 (2025). https://doi.org/10.1038/s41596-024-01135-3
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41596-024-01135-3