Abstract
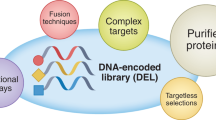
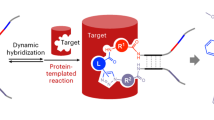
Solid-phase DNA-encoded library (DEL) synthesis is a next-generation drug discovery technology with powerful activity-based and cellular lead identification capabilities. Solid-phase DELs combine the one-bead–one-compound approach with DNA encoding to furnish beads that display multiple copies of photocleavable library members and DNA encoding tags. Sequential chemical synthesis and enzymatic DNA ligation reactions yield an encoded library in which individual library members are physically isolable, enabling various high-throughput screening modalities. This advancement from on-DNA synthesis, in which small molecules are directly attached to their DNA-encoding tags, decouples the library member from the steric bulk of the DNA tag, which prevents biased binding to a target. Here we provide step-by-step instructions for solid-phase DEL synthesis, incorporating all of our most recent quality control innovations to ensure robust library production. The protocol begins with on-bead synthesis of a linker containing a spectroscopic handle for chromatographic analysis, an ionization enhancer for mass spectrometry and an alkyne for installation of DNA encoding sites via copper-catalyzed azide-alkyne cycloaddition click chemistry. Coupling of a photocleavable linker before library synthesis enables compound liberation from the bead for activity-based screening. Powerful combinatorial split-and-pool parallel synthesis tactics transform modest collections of small-molecule building blocks into large DELs of all possible building block combinations. Post synthesis, decoding and mass analysis of single DEL beads as well as whole-library deep sequencing provides rigorous chemical and bioinformatic quality control and establishes suitability for screening. The solid-phase chemistry is highly accessible: expertise in chemical synthesis is not necessary and solid-phase synthesis apparatus is routinely available in molecular biology laboratories. This procedure requires ~1 month to complete.
Key points
-
This solid-phase DNA-encoded library synthesis protocol uses combinatorial split-and-pool synthesis to generate diverse, photocleavable chemical compounds encoded by DNA tags.
-
Solid-phase chemistry using a photocleavable linker allows off-DNA release and evaluation of small-molecule library members, enabling activity-based screening for DNA-encoded libraries.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Example NGS decoding scripts and raw data files accompanying figures within this manuscript can be found at Example NGS Decoding script via Figshare at https://figshare.com/s/425bb12ef2b5f20bb856 (ref. 45). The raw data files can be found via Figshare at https://figshare.com/s/5453000e506029b5298c (ref. 46). Source data are provided with this paper.
References
Carter, A. J. et al. Target 2035: probing the human proteome. Drug Discov. Today 24, 2111–2115 (2019).
Zhang, Y. & Clark, M. A. Design concepts for DNA-encoded library synthesis. Bioorg. Med. Chem. 41, 116189 (2021).
Henley, M. J. & Koehler, A. N. Advances in targeting ‘undruggable’ transcription factors with small molecules. Nat. Rev. Drug Discov. 20, 669–688 (2021).
Coleman, N. & Rodon, J. Taking aim at the undruggable. Am. Soc. Clin. Oncol. Educ. Book 41, e145–e152 (2021).
Dixit, A., Barhoosh, H. & Paegel, B. M. Translating the genome into drugs. Acc. Chem. Res. 56, 489–499 (2023).
Lam, K. S., Lebl, M. & Krchňák, V. The ‘One-Bead-One-Compound’ combinatorial library method. Chem. Rev. 97, 411–448 (1997).
Brenner, S. & Lerner, R. A. Encoded combinatorial chemistry. Proc. Natl. Acad. Sci. USA 89, 5381–5383 (1992).
Melkko, S., Scheuermann, J., Dumelin, C. E. & Neri, D. Encoded self-assembling chemical libraries. Nat. Biotechnol. 22, 568–574 (2004).
Clark, M. A. et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat. Chem. Biol. 5, 647–654 (2009).
Mannocci, L. et al. High-throughput sequencing allows the identification of binding molecules isolated from DNA-encoded chemical libraries. Proc. Natl. Acad. Sci. USA 105, 17670–17675 (2008).
Harris, P. A. et al. DNA-encoded library screening identifies benzo[b][1,4]oxazepin-4-ones as highly potent and monoselective receptor interacting protein 1 kinase inhibitors. J. Med. Chem. 59, 2163–2178 (2016).
Cuozzo, J. W. et al. Novel autotaxin inhibitor for the treatment of idiopathic pulmonary fibrosis: A clinical candidate discovered using DNA-encoded chemistry. J. Med. Chem. 63, 7840–7856 (2020).
Kung, P.-P. et al. Characterization of specific N‑α-acetyltransferase 50 (Naa50) inhibitors identified using a DNA encoded library. ACS Med. Chem. Lett. 11, 1175–1184 (2020).
Ding, Y. et al. Discovery of soluble epoxide hydrolase inhibitors through DNA-encoded library technology (ELT). Bioorg. Med. Chem. 41, 116216 (2021).
Machutta, C. A. et al. Prioritizing multiple therapeutic targets in parallel using automated DNA-encoded library screening. Nat. Commun. 8, 16081 (2017).
Cai, B. et al. Selection of DNA-encoded libraries to protein targets within and on living cells. J. Am. Chem. Soc. 141, 17057–17061 (2019).
Litovchick, A. et al. Novel nucleic acid binding small molecules discovered using DNA-encoded chemistry. Molecules 24, 2026 (2019).
Huang, Y. et al. Selection of DNA-encoded chemical libraries against endogenous membrane proteins on live cells. Nat. Chem. 13, 77–88 (2021).
Satz, A. L., Kuai, L. & Peng, X. Selections and screenings of DNA-encoded chemical libraries against enzyme and cellular targets. Bioorg. Med. Chem. Lett. 39, 127851 (2021).
Benhamou, R. I. et al. DNA-encoded library versus RNA-encoded library selection enables design of an oncogenic noncoding RNA inhibitor. Proc. Natl. Acad. Sci. USA 119, e2114971119 (2022).
Kolb, H. C., Finn, M. G. & Sharpless, K. B. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 40, 2004–2021 (2001).
MacConnell, A. B., McEnaney, P. J., Cavett, V. J. & Paegel, B. M. DNA-encoded solid-phase synthesis: encoding language design and complex oligomer library synthesis. ACS Comb. Sci. 17, 518–534 (2015).
Malone, M. L. & Paegel, B. M. What is a ‘DNA-compatible’ reaction? ACS Comb. Sci. 18, 182–187 (2016).
Price, A. K., MacConnell, A. B. & Paegel, B. M. hνSABR: photochemical dose–response bead screening in droplets. Anal. Chem. 88, 2904–2911 (2016).
MacConnell, A. B., Price, A. K. & Paegel, B. M. An integrated microfluidic processor for DNA-encoded combinatorial library functional screening. ACS Comb. Sci. 19, 181–192 (2017).
Cochrane, W. G. et al. Activity-based DNA-encoded library screening. ACS Comb. Sci. 21, 425–435 (2019).
Fitzgerald, P. R., Cochrane, W. G. & Paegel, B. M. Dose–response activity-based DNA-encoded library screening. ACS Med. Chem. Lett. 14, 1295–1303 (2023).
Barhoosh, H. et al. Activity-based DNA-encoded library screening for selective inhibitors of eukaryotic translation. ACS Cent. Sci. 10, 1960–1968 (2024).
Hackler, A. L., FitzGerald, F. G., Dang, V. Q., Satz, A. L. & Paegel, B. M. Off-DNA DNA-encoded library affinity screening. ACS Comb. Sci. 22, 25–34 (2020).
Hu, J. et al. Liposomal permeation assay for droplet-scale pharmacokinetic screening. J. Med. Chem. 66, 6288–6296 (2023).
Mendes, K. R. et al. High-throughput identification of DNA-encoded IgG ligands that distinguish active and latent Mycobacterium tuberculosis infections. ACS Chem. Biol. 12, 234–243 (2017).
Gibaut, Q. M. R. et al. Study of an RNA-focused DNA-encoded library informs design of a degrader of a r(CUG) repeat expansion. J. Am. Chem. Soc. 144, 21972–21979 (2022).
Cochrane, W. G., Fitzgerald, P. R. & Paegel, B. M. Antibacterial discovery via phenotypic DNA-encoded library screening. ACS Chem. Biol. 16, 2752–2756 (2021).
McCloskey, K. et al. Machine learning on DNA-encoded libraries: a new paradigm for hit finding. J. Med. Chem. 63, 8857–8866 (2020).
Lim, K. S. et al. Machine learning on DNA-encoded library count data using an uncertainty-aware probabilistic loss function. J. Chem. Inf. Model. 62, 2316–2331 (2022).
Hou, R., Xie, C., Gui, Y., Li, G. & Li, X. Machine-learning-based data analysis method for cell-based selection of DNA-encoded libraries. ACS Omega 8, 19057–19071 (2023).
Ratnayake, A. S. et al. A solution phase platform to characterize chemical reaction compatibility with DNA-encoded chemical library synthesis. ACS Comb. Sci. 21, 650–655 (2019).
Satz, A. L. et al. DNA compatible multistep synthesis and applications to DNA encoded libraries. Bioconjug. Chem. 26, 1623–1632 (2015).
Fitzgerald, P. R. & Paegel, B. M. DNA-encoded chemistry: drug discovery from a few good reactions. Chem. Rev. 121, 7155–7177 (2021).
Fitzgerald, P. R., Dixit, A., Zhang, C., Mobley, D. L. & Paegel, B. M. Building block-centric approach to DNA-encoded library design. J. Chem. Inf. Model. 64, 4661–4672 (2024).
Kivioja, T. et al. Counting absolute number of molecules using unique molecular identifiers. Nat. Methods https://doi.org/10.1038/npre.2011.5903.1 (2011).
MacConnell, A. B. & Paegel, B. M. Poisson statistics of combinatorial library sampling predict false discovery rates of screening. ACS Comb. Sci. 19, 524–532 (2017).
Mikkelsen, R. J. T., Grier, K. E., Mortensen, K. T., Nielsen, T. E. & Qvortrup, K. Photolabile linkers for solid-phase synthesis. ACS Comb. Sci. 20, 377–399 (2018).
ChaAn, T. R., Hilgraf, R., Sharpless, K. B. & Fokin, V. V. Polytriazoles as copper(I)‐stabilizing ligands in catalysis. Organic Lett. 6, 2853–2855 (2004).
Dixit, A., Cavett, V. & Paegel, B. M. NGS decoding example script (Python). Figshare https://figshare.com/s/425bb12ef2b5f20bb856 (2024).
Dixit, A. & Paegel, B. M. Raw data. Figshare https://figshare.com/s/5453000e506029b5298c (2024).
Acknowledgements
This work was supported by a grant award from the National Institutes of Health (grant no. GM140890) to B.M.P.
Author information
Authors and Affiliations
Contributions
A.D. collected and analyzed data and prepared a preliminary draft of the manuscript with guidance from B.M.P. All authors read, edited and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Xiaojie Lu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key references
Barhoosh, H. et al. ACS Cent Sci. 10, 1960–1968 (2024): https://doi.org/10.1021/acscentsci.4c01218
Fitzgerald, P. et al. ACS Med. Chem. Lett. 14, 1295–1303 (2023): https://doi.org/10.1021/acsmedchemlett.3c00159
Hackler, A. et al. ACS Comb Sci. 22, 25–34 (2020): https://doi.org/10.1021/acscombsci.9b00153
Supplementary information
Supplementary Information
Supplementary Tables 1–6 and Figs. 1–7.
Source data
Source Data Fig. 4
Source plots for chromatography plots and mass spectral data.
Source Data Fig. 6
Unprocessed PAGE gels and source MALDI-TOF MS data for figure. Available in Supplementary Information as well.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dixit, A., Paegel, B.M. Solid-phase DNA-encoded library synthesis: a master builder’s instructions. Nat Protoc (2025). https://doi.org/10.1038/s41596-025-01190-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-025-01190-4