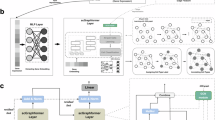
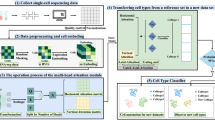
Abstract
Single-cell research faces challenges in accurately annotating cell types at high resolution, especially when dealing with large-scale datasets and rare cell populations. To address this, foundation models such as single-cell generative pretrained transformer (scGPT) offer flexible, scalable solutions by leveraging transformer-based architectures. Here we provide a comprehensive guide to fine-tuning scGPT for cell-type classification in single-cell RNA sequencing data. We demonstrate how to fine-tune scGPT on a custom retina dataset, highlighting the model’s efficiency in handling complex data and improving annotation accuracy achieving 99.5% F1-score. This protocol automates key steps, including data preprocessing, model fine-tuning and evaluation. This protocol enables researchers to efficiently deploy scGPT for their own datasets. The provided tools, including a command-line script and Jupyter Notebook, simplify the customization and exploration of the model, proposing an accessible workflow for users with minimal Python and Linux knowledge. The protocol offers an off-the-shell solution of high-precision cell-type annotation using scGPT for researchers with intermediate bioinformatics. The source code and example datasets are publicly available on GitHub and Zenodo.
Key points
-
This protocol provides the instructions to automating key steps, including data preprocessing, model fine-tuning and evaluation for single-cell generative pretrained transformer using Python function wrappers within computing clusters and Jupyter notebooks.
-
The single-cell generative pretrained transformer protocol provides a structured framework for single-cell analysis using the pretrained foundation model and serves as an alternative to methods such as Seurat, scPred, scArches or Geneformer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The example snRNA-seq dataset used in this protocol are available via Zenodo at https://doi.org/10.5281/zenodo.14648190 (ref. 28).
Code availability
The code for this protocol is available via GitHub at https://github.com/RCHENLAB/scGPT_fineTune_protocol. A detailed Jupyter Notebook is also provided for use with both Google Colab and JupyterLab.
References
Cui, H. et al. scGPT: toward building a foundation model for single-cell multi-omics using generative AI. Nat. Methods 21, 1470–1480 (2024).
Theodoris, C. V. et al. Transfer learning enables predictions in network biology. Nature 618, 616–624 (2023).
Rosen, Y. et al. Universal cell embeddings: a foundation model for cell biology. Preprint at bioRxiv https://doi.org/10.1101/2023.11.28.568918 (2023).
Lotfollahi, M. et al. Mapping single-cell data to reference atlases by transfer learning. Nat. Biotechnol. 40, 121–130 (2022).
Yang, F. et al. scBERT as a large-scale pretrained deep language model for cell type annotation of single-cell RNA-seq data. Nat. Mach. Intell. 4, 852–866 (2022).
Alquicira-Hernandez, J., Sathe, A., Ji, H. P., Nguyen, Q. & Powell, J. E. scPred: accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol. 20, 264 (2019).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Li, J. et al. Integrated multi-omics single cell atlas of the human retina. Preprint at bioRxiv https://doi.org/10.1101/2023.11.07.566105 (2023).
Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
Xu, C. et al. Probabilistic harmonization and annotation of single‐cell transcriptomics data with deep generative models. Mol. Syst. Biol. 17, e9620 (2021).
Welch, J. D. et al. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 177, 1873–1887 (2019).
Bian, H. et al. in Research in Computational Molecular Biology (ed. Ma, J.) 479–482 (Springer Nature, 2024).
Jiao, L. et al. scTransSort: transformers for intelligent annotation of cell types by gene embeddings. Biomolecules 13, 611 (2023).
Chen, J. et al. Transformer for one stop interpretable cell type annotation. Nat. Commun. 14, 223 (2023).
Clarke, Z. A. et al. Tutorial: guidelines for annotating single-cell transcriptomic maps using automated and manual methods. Nat. Protoc. 16, 2749–2764 (2021).
Cheng, C., Chen, W., Jin, H. & Chen, X. A review of single-cell RNA-seq annotation, integration, and cell–cell communication. Cells 12, 1970 (2023).
Yu, X., Xu, X., Zhang, J. & Li, X. Batch alignment of single-cell transcriptomics data using deep metric learning. Nat. Commun. 14, 960 (2023).
Haghverdi, L., Lun, A. T. L., Morgan, M. D. & Marioni, J. C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427 (2018).
Nguyen, H. C. T., Baik, B., Yoon, S., Park, T. & Nam, D. Benchmarking integration of single-cell differential expression. Nat. Commun. 14, 1570 (2023).
Vaswani, A. et al. in Advances in Neural Information Processing Systems Vol. 30 (Guyon, I. et al.) 6000–6010 (Curran Associates, 2017).
Boser, B. E., Guyon, I. M. & Vapnik, V. N. A training algorithm for optimal margin classifiers. In Proc. Fifth Annual Workshop on Computational Learning Theory 144–152 (Association for Computing Machinery, 1992).
Cortes, C. & Vapnik, V. Support-vector networks. Mach. Learn. 20, 273–297 (1995).
Biewald, L. Weights & Biases: the AI developer platform. Weights & Biases https://wandb.ai/site (2020).
Hahn, J. et al. Evolution of neuronal cell classes and types in the vertebrate retina. Nature 624, 415–424 (2023).
Wang, S. K. et al. Single-cell multiome of the human retina and deep learning nominate causal variants in complex eye diseases. Cell Genomics 2, 100164 (2022).
Lukowski, S. W. et al. A single‐cell transcriptome atlas of the adult human retina. EMBO J. 38, e100811 (2019).
Ding, S. et al. scGPT: end-to-end protocol for fine-tuned retina cell type annotation. Zenodo https://doi.org/10.5281/zenodo.14648190 (2025).
Chawla, N. V., Bowyer, K. W., Hall, L. O. & Kegelmeyer, W. P. SMOTE: synthetic minority over-sampling technique. J. Artif. Intell. Res. 16, 321–357 (2002).
Chen, J. et al. Deep transfer learning of cancer drug responses by integrating bulk and single-cell RNA-seq data. Nat. Commun. 13, 6494 (2022).
Khan, S. A. et al. Reusability report: learning the transcriptional grammar in single-cell RNA-sequencing data using transformers. Nat. Mach. Intell. 5, 1437–1446 (2023).
Cheng, Y., Fan, X., Zhang, J. & Li, Y. A scalable sparse neural network framework for rare cell type annotation of single-cell transcriptome data. Commun. Biol. 6, 1–13 (2023).
Acknowledgements
This work was supported by Chan-Zuckerburg Foundation (grant nos. CZF2021-237885 and CZF2019-002425 to R.C). The authors acknowledge support to the Gavin Herbert Eye Institute at the University of California, Irvine from an unrestricted grant from Research to Prevent Blindness and from NIH (grant no. P30 EY034070).
Author information
Authors and Affiliations
Contributions
S.D. and H.C. developed the protocol. R.L. contributed to hyperparameter-tuning and code quality testing. J.L. performed data preparation and data analysis. R.C. supervised the biological aspects and data analysis. B.W. supervised the fine-tuning procedure. S.D., J.L. and R.L. prepared the manuscript. All authors critically reviewed the manuscript and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
All authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key reference
Cui, H. et al. Nat Methods 21, 1470–1480 (2024): https://doi.org/10.1038/s41592-024-02201-0
Supplementary information
Supplementary Information
Supplementary Figs. 1–6.
Supplementary Tables 1–4
Supplementary Table 1. Available variables in the preprocess pipeline. Required variables are marked with ‘[REQUIRED]’, while others are optional. Supplementary Table 2. Available variables in the fine-tuning pipeline. Required variables are marked with ‘[REQUIRED]’, while others are optional. Supplementary Table 3. Available variables in the inference pipeline. Required variables are marked with ‘[REQUIRED]’, while others are optional. Supplementary Table 4. Available variables in the zero-shot inference pipeline. Required variables are marked with ‘[REQUIRED]’, while others are optional.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ding, S., Li, J., Luo, R. et al. scGPT: end-to-end protocol for fine-tuned retinal cell type annotation. Nat Protoc (2025). https://doi.org/10.1038/s41596-025-01220-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41596-025-01220-1