Abstract
Diatoms are microalgae that live in marine and freshwater environments and are responsible for about 20% of the world’s carbon fixation. Population dynamics of these cells is finely regulated by intricate signal transduction systems, in which oxylipins are thought to play a relevant role. These are oxygenated fatty acids whose biosynthesis is initiated by a lipoxygenase enzyme (LOX) and are widely distributed in all phyla, including diatoms. Here, we present a de novo transcriptome obtained from the RNA-seq performed in the diatom species Pseudo-nitzschia arenysensis, using both a wild-type and a LOX-silenced strain, which will represent a reliable reference for comparative analyses within the Pseudo-nitzschia genus and at a broader taxonomic scale. Moreover, the RNA-seq data can be interrogated to go deeper into the oxylipins metabolic pathways.
Similar content being viewed by others
Background & Summary
Diatoms are a group of very diverse photosynthetic microorganisms that evolved a broad range of adaptive strategies allowing them to prosper under a wide variety of temperature, light, and nutrient conditions1. Thanks to these characteristics, they populate almost all aquatic and wet environments, contributing to ca. 20% of global carbon fixation1,2; additionally, as important primary producers, they form the basis of aquatic food webs. The genus Pseudo-nitzschia is one of the most common genera of diatoms, comprising about 60 worldwide distributed species, among which 26 species produce the neurotoxin domoic acid and are responsible for harmful algal blooms3. Among the species of this group, P. arenysensis4 is a non-toxic species that regularly blooms in coastal and oceanic waters5; its life cycle has been well described6 and the transcriptome and an optimized transformation protocol are already available7,8,9. All these characteristics made this species a good model for functional and comparative genomic studies.
The high rate of biodiversity characterizing diatoms makes them producers of high-value bioactive compounds, whose identification could be exploitable by the biotechnology industry10,11, such as oxylipins, oxygenated fatty acids involved in the reduction of grazing pressure12,13, in the chemical communications regulating phytoplankton dynamics14,15,16 and the interactions with bacteria17. Lipoxygenase enzymes (LOXs), a group of nonheme iron-containing dioxygenases, are responsible for the biosynthesis of these metabolites18,19,20,21,22,23. Distinct oxylipins are produced by different LOXs, depending on the polyunsaturated fatty acid (PUFA) used as substrate and the position on the carbon backbone where oxygen (O2) is added18.
Pseudo-nitzschia members produce a wide range of species-specific oxylipins24, suggesting differentiation of LOX enzymes to ensure to these metabolites a species-specific mediator role in the plankton community24,25. To investigate the ecological roles of these secondary metabolites, studies that examine their structure and biosynthesis should be paralleled with functional studies on the enzyme-coding genes involved in their synthesis. In recent work, we explored the role of LOX in P. arenysensis (PaLOX)26, a diatom known to synthesize oxylipins throughout both 12- and 15S-LOX pathways14,16,24. Taking advantage of sequence information from the Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP)7 we discovered that a unique LOX transcript is present in this species and generated mutants in which this gene was silenced26.
Here, we describe the de novo transcriptome assembly of a LOX-interfered P. arenysensis clone and of the corresponding wild-type strain, which captures two different conditions with respect to that pictured by the P. arenysensis transcriptome sequenced within the MMETSP7. The availability of this new transcriptome, built by exploiting new datasets and upgraded pipelines and software, is useful because it allows to enrich the information of previously assembled transcriptomes by adding or updating missing or badly annotated transcripts27. Furthermore, to elucidate the molecular mechanisms and the gene networks underlying diatoms adaptability, the “-omics” technologies have spread considerably, and the genome of an ever-increasing number of diatom species, among which P. arenysensis, is being sequenced. Within this context, the 12 RNA-seq datasets released with this paper, together with previously generated datasets for the same species, will aid in building accurate gene models and will allow to enrich the species gene expression atlas. In addition, the integration of the information deriving from the genome sequence analysis, the transcriptome of different strains, the gene expression profiles, and the comparison with similar data from the other diatom species will shed light on key genes mediating adaptation across the global ocean. Since the timing for the genome release is still unknown, the present transcriptome represents a resource allowing more accurate comparative genomic analyses within the Pseudo-nitzschia genus and at a broader taxonomic scale.
Moreover, we already demonstrated that the oxylipin reduction in P. arenysensis results in growth impairment of the interfered cells compared to the WT26, and the RNA-seq data can be interrogated to reveal the underlying perturbated pathways. Finally, the identification of P. arenysensis molecular mechanisms tuning the oxylipin-mediated cell growth regulation can be used as a guide in determining a correlation between diatom cell growth/density and oxylipin concentrations in other diatom species.
Methods
Culture growth condition and RNA preparation
The wild-type SV6 strain of P. arenysensis was obtained from crosses performed in the laboratory from strains isolated at the Mare Chiara LTER station in the Gulf of Naples. The LOX-interfered P. arenysensis Int11 sample, derived from the biolistic transformation of SV6 cells, has been described in Sabatino et al.26. Cultures were grown in seawater enriched with F/2 nutrient28 and incubated at 18 °C under white light at approximately 70 μmol photons m−2 s−1 and 12:12 h dark:light cycle. For the growth curves, fresh diatom cultures were inoculated at a start cell density of 5000 cells mL−1, grown under the above-mentioned conditions. The growth was monitored daily by cell counting under a Zeiss inverted microscope and using Malassez chambers of 100 μL capacity. Each curve was performed in triplicate. Cells were collected at two time points of the growth curve, the stationary phase (T10) and the senescence phase (T12) (Fig. 1 and Supplementary Fig. 1). The RNA extraction was performed following the protocol detailed in Amato et al.29. Specifically, mutated and wild-type P. arenysensis cultures were filtered onto RAWP Millipore cellulose membrane filters (Mf-Millipore RAWP04700, 1.2 µm porosity) and washed with MilliQ water. Filters were put into 2 ml Eppendorf tubes filled with Roche TriPure® isolation reagent (Merck KGaA, Darmstadt, Germany), snap-frozen in liquid nitrogen, and stored at −80 °C until use. TriPure®-soaked filters were thawed at room temperature and RNA extraction was conducted according to the manufacturer’s instructions. RNA samples were DNAse-treated (TURBO DNA-free™ Kit, Waltham, Massachusetts, USA) to get rid of genomic DNA contamination and purified with Qiagen RNeasy Mini Kit (Qiagen, Hilden, Germany). PCRs were performed using the RNA as template to verify absence of genomic DNA contamination. Samples were stored at −80 °C until use.
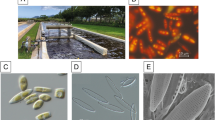
Diagram illustrating the experimental design. (a) Western blot analysis of the P. arenysensis LOX protein, PaLOX, performed on the wild-type (SV6) and three silenced samples (Int4, Int11, Int20); the beta subunit of the ATP synthase, Atpb, was used as an internal control. LOX antibodies bind to a protein of c. 79 kDa, AtpB antibodies bind to a protein of c. 54 kDa; (b) Growth curve of the transformant Int11 (dotted line/closed square) with its control SV6 (solid line/open square). Red arrows indicate sampling points for RNA-seq, day 10 (T10) and day 12 (T12). At those time points, the growth of the transformant strain compared to the control was reduced of approximately 11% and 50%, respectively. Mean values obtained from three biological replicates and SD are presented.
Quality control of total RNA samples, library preparation and sequencing
RNA samples were analyzed on an Agilent 2100 Bioanalyzer platform (Agilent Technologies 5301 Stevens Creek Blvd. Santa Clara, California 95051 USA) to assess integrity, on a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA) to assess purity, and quantified with a Qubit fluorometer (Thermo Fisher Scientific Inc.). 300 ng of RNA from each sample per time point was used to produce libraries with the Illumina TruSeq original protocol, with a bead-based poly-A capture approach, and sequenced on an Illumina HiSeq 2000 (Single End 50 bp reads) at the GeneCore facility of the European Molecular Biology Laboratory (EMBL).
Reads quality check, transcriptome assembly and gene annotation
The quality check of the raw reads was performed using FASTQC30. A trimming step was carried out by Trimmomatic31, setting a minimum lentgh of the reads to 30 bp and using the “ILLUMINACLIP” parameter to remove TruSeq Adapters. All the cleaned reads were assembled into transcript sequences using Trinity v.2.1532 with in silico read normalization, setting the -min_kmer_cov parameter at 2. The clustering of the transcriptome was performed using the CD-hit software v. 4.6.833 with 90% identity threshold in order to remove transcriptome redundancy. Since we obtained a partial assembly of the lipoxygenase (LOX) transcript, the complete assembly of the LOX was performed by Spades v.3.15.434, setting the k-mer size to 23 and using the LOX sequence obtained by previous experiments26 as the object of the “–trusted-contigs” parameter. 695 sequences from bacteria or viruses, identified via the Transcriptome Shotgun Assembly (TSA) web portal at NCBI35, were filtered out. Moreover, one transcript sequence with less than 200 bases length was also removed. The completeness of the transcriptome was evaluated by using Busco v.5.7.036, setting stramenopiles as the lineage of search. The whole transcriptome was aligned with BLASTx software37 versus the Uniprot/SwissProt database38 (downloaded in September 2022), setting the e-value threshold to 1e−3, and retrieving the best hit for each assembled transcript.
Expression analysis
Cleaned reads were mapped on the assembled transcriptome with bowtie v.2.3.4.139. Reads counts for each replicate were performed using the eXpress software40. The hierarchical clustering dendrogram was obtained with the WGCNA package in R41, setting an “average” distance parameter in the hclust function. The principal component analysis (PCoA) was performed by Scikit-learn module42 in Python programming language43, and plotted by using seaborn44.
Data Records
All the RNA-Seq raw reads generated in this project were deposited in the NCBI Sequence Read Archive database with identifier SRP40888045, under project identification number PRJNA903772. The de novo transcriptome assembly resource (final transcriptome) is available in the NCBI Transcriptome Shotgun Assembly with accession GKNO0000000046 and in the Zenodo entry, where we also added the annotated genes file as XLS file47.
Technical Validation
Quality control
RNA-seq experiments45 were performed on two strains of P. arenysensis; in detail, six samples of a wild-type strain culture and six samples of an interfered strain culture were collected (two time points in triplicate per strain). The pre-trimming FASTQC step enabled us to identify the TruSeq adapters to be removed in the next cleaning step. The trimming procedure removed all the reads shorter than 30 bp, together with the identified TruSeq adapters. The post-trimming FASTQC step allowed us to check that all the trimmed reads retained a minimum quality PHRED score of 30.
Transcriptome assembly and annotation
Trimmed reads have been assembled into a transcriptome46 with a de novo approach. The assembled transcriptome accounted for a total of 31758 transcripts (Table 1), with a mean GC content of 46.01%, an average and median contig length of 925.58 and 681 bp, respectively, and a N50 of 1417 bp (Table 1). The assembled transcriptome was subjected to a clustering procedure in order to remove redundancy. Moreover, a step to filter out sequences from bacteria or viruses and sequences shorter than 200 bases in length was performed, removing 696 sequences. After this step, the final transcriptome accounted for a total of 27784 transcripts, in the same range of the MMETSP P. arenysensis transcriptome7 (MMETSP0329). The procedure of functional annotation enabled the functional classification of 8857 transcript sequences (31.9% of the total transcriptome, Table 1).
A summary of the number of reads obtained from the sequencing step and their mapping on the assembled transcriptome is shown in Table 2.
The BUSCO analysis revealed that the transcriptome has 91 complete stramenopiles BUSCO genes over 100 total genes (89 single-copy and 2 duplicated genes), with only 4% completely missing (Fig. 2).
Preliminary expression results
The sample clustering of the expression levels of each replicate revealed a sharp distinction between wild-type and knock-down samples (Fig. 3a). The principal component analysis revealed a similar clear discrimination between wild-type and interfered samples, together with a well-defined separation within the knock-down samples between the stationary phase (T10) replicates and the senescence phase (T12) replicates (Fig. 3b).
RNA-seq samples similarity analysis. (a) Dendrogram of hierarchical sample clustering, wild-type samples are highlighted by the pink box, and knock-down samples are highlighted by the green box. (b) Principal component analysis of RNA-seq samples; different sample categories are indicated according to the legend.
Usage Notes
The transcriptome46 presented here represents an alternative to the MMETSP7 P. arenysensis transcriptome, and together with it, it allows to extract information from two different strains from the same geographical location (B593 and SV6) and different physiological conditions (SV6 wild-type and SV6 PaLOX-silenced). Compared to BUSCO statistics of the MMETSP transcriptome (60 complete single-copy genes, 10 missing genes), the transcriptome we presented improved the number of complete single-copy genes (89) and reduced the number of missing genes (4). A summary of the transcriptome statistics is shown in Supplementary Table 1 (S1). All these resources will be useful for a high-quality gene model prediction in the perspective of sequencing the P. arenysensis genome48, which in turn will be extremely useful for comparative genomics studies when other diatom genomes are released, such as those planned within the “100 Diatoms Genomes Project” at the Joint Genome Institute (JGI)49. Moreover, our data could be an important reference in large-scale metagenomic and metatranscriptomic data analyses of eukaryotic plankton in the open ocean5 and coastal ecosystems, such as those collected within the TARA Oceans50 and TREC (https://www.embl.org/about/info/trec/) expeditions, respectively, or the augmented observatory NEREA (https://www.nerea-observatory.org/). Finally, transcriptome data from the LOX-interfered P. arenysensis strain45 provide a foundation for future detailed studies on the oxylipin-mediated cell signaling pathways in this and in other diatom species, while the availability of different P. arenysensis RNA-seq datasets could also be useful to uncover single nucleotide polymorphisms (SNPs) in the coding regions of the genome.
Code availability
The article includes a list of software programs, such as de novo transcriptome assembly, pre- and post-assembly procedures, and transcriptome annotation, all of which are specified alongside their respective versions within the Methods section. If specific parameter details are not provided, the programs were used with their default settings.
References
Armbrust, E. V. The life of diatoms in the world’s oceans. Nat. 459, 185–192 (2009). 2009 4597244.
Field, C. B., Behrenfeld, M. J., Randerson, J. T. & Falkowski, P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science (80-) 281, 237–240 (1998).
Bates, S. S., Hubbard, K. A., Lundholm, N., Montresor, M. & Leaw, C. P. Pseudo-nitzschia, Nitzschia, and domoic acid: New research since 2011. Harmful Algae 79, 3–43 (2018).
Quijano-Scheggia, S. I. et al. Morphology, physiology, molecular phylogeny and sexual compatibility of the cryptic Pseudo-nitzschia delicatissima complex (Bacillariophyta), including the description of P. arenysensis sp. nov. Phycologia 48, 492–509 (2009).
Trainer, V. L. et al. Pseudo-nitzschia physiological ecology, phylogeny, toxicity, monitoring and impacts on ecosystem health. Harmful Algae 14, 271–300 (2012).
Amato, A., Orsini, L., D’Alelio, D. & Montresor, M. Life cycle, size reduction patterns, and ultrastructure of the pennate planktonic diatom Pseudo-nitzschia delicatissima (bacillariophyceae). J. Phycol. 41, 542–556 (2005).
Keeling, P. J. et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the Functional Diversity of Eukaryotic Life in the Oceans through Transcriptome Sequencing. PLoS Biol. 12, e1001889 (2014).
Sabatino, V. et al. Establishment of Genetic Transformation in the Sexually Reproducing Diatoms Pseudo-nitzschia multistriata and Pseudo-nitzschia arenysensis and Inheritance of the Transgene. Mar. Biotechnol. 17, 452–462 (2015).
Di Dato, V. et al. Transcriptome sequencing of three Pseudo-nitzschia species reveals comparable gene sets and the presence of Nitric Oxide Synthase genes in diatoms. Sci. Rep. 5 (2015).
Eltanahy, E. & Torky, A. CHAPTER 1:Microalgae as Cell Factories: Food and Feed-grade High-value Metabolites. Microalgal Biotechnol. 1–35, https://doi.org/10.1039/9781839162473-00001 (2021).
Balasubramaniam, V., Gunasegavan, R. D. N., Mustar, S., Lee, J. C. & Noh, M. F. M. Isolation of Industrial Important Bioactive Compounds from Microalgae. Mol. 26, 943 (2021).
Miralto, A. et al. The insidious effect of diatoms on copepod reproduction. Nature 402, 173–176 (1999).
Ianora, A., Poulet, S. A. & Miralto, A. The effects of diatoms on copepod reproduction: A review. Phycologia 42, 351–363 (2003).
Fontana, A. et al. LOX-induced lipid peroxidation mechanism responsible for the detrimental effect of marine diatoms on zooplankton grazers. ChemBioChem 8, 1810–1818 (2007).
Barreiro, A. et al. Diatom induction of reproductive failure in copepods: The effect of PUAs versus non volatile oxylipins. J. Exp. Mar. Bio. Ecol. 401, 13–19 (2011).
D’Ippolito, G. et al. 15S-Lipoxygenase metabolism in the marine diatom Pseudo-nitzschia delicatissima. New Phytol. 183, 1064–1071 (2009).
Meyer, N., Rettner, J., Werner, M., Werz, O. & Pohnert, G. Algal Oxylipins Mediate the Resistance of Diatoms against Algicidal Bacteria. Mar. Drugs 16, 486 (2018).
Brash, A. R. Lipoxygenases: Occurrence, functions, catalysis, and acquisition of substrate. Journal of Biological Chemistry 274, 23679–23682 (1999).
D’Ippolito, G. et al. Production of octadienal in the marine diatom Skeletonema costatum. Org. Lett. 5, 885–887 (2003).
D’Ippolito, G. et al. New C16 fatty-acid-based oxylipin pathway in the marine diatom Thalassiosira rotula. Org. Biomol. Chem. 3, 4065–4070 (2005).
Lion, U. et al. Phospholipases and galactolipases trigger oxylipin-mediated wound-activated defence in the red alga Gracilaria chilensis against epiphytes. ChemBioChem 7, 457–462 (2006).
Andreou, A., Brodhun, F. & Feussner, I. Biosynthesis of oxylipins in non-mammals. Progress in Lipid Research 48, 148–170 (2009).
Bonaventure, G. Lipases and the biosynthesis of free oxylipins in plants. Plant Signal. Behav. 9, 9–12 (2014).
Lamari, N. et al. Specificity of Lipoxygenase Pathways Supports Species Delineation in the Marine Diatom Genus Pseudo-nitzschia. PLoS One 8, e73281 (2013).
Jenke-Kodama, H., Müller, R. & Dittmann, E. Evolutionary mechanisms underlying secondary metabolite diversity. Progress in Drug Research 65, 120–140 (2008).
Sabatino, V. et al. Silencing of a Pseudo-nitzschia arenysensis lipoxygenase transcript leads to reduced oxylipin production and impaired growth. New Phytol. https://doi.org/10.1111/NPH.17739 (2021).
Martin, J. A. & Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 12, 671–682 (2011). 2011 1210.
Guillard, R. R. L. Culture of Phytoplankton for Feeding Marine Invertebrates. in Culture of Marine Invertebrate Animals 29–60. https://doi.org/10.1007/978-1-4615-8714-9_3 (Springer US, 1975).
Amato, A. et al. Grazer-induced transcriptomic and metabolomic response of the chain-forming diatom Skeletonema marinoi. ISME J. 12, 1594–1604 (2018).
Simon Andrews Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data. Soil 5, 47–81 (2020).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Sayers, E. W. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 49, D10 (2021).
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A. & Zdobnov, E. M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 38, 4647 (2021).
Camacho, C. et al. BLAST+: Architecture and applications. BMC Bioinformatics 10 (2009).
Boutet, E. et al. UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods Mol. Biol. 1374, 23–54 (2016).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Roberts, A., Trapnell, C., Donaghey, J., Rinn, J. L. & Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12 (2011).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9 (2008).
Pedregosa, F. et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Van Rossum, G. & Drake, F. L. Python 3 Reference Manual; CreateSpace. Scotts Val. CA 242 (2009).
Waskom, M. L. seaborn: statistical data visualization. J. Open Source Softw. 6, 3021 (2021).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP408880 (2023).
Marotta, P., Sabatino, V., Ambrosino, L., Miralto, M. & Ferrante, M. I. Pseudo-nitzschia arenysensis, transcriptome shotgun assembly. GenBank https://identifiers.org/ncbi/insdc:GKNO00000000 (2023).
Sabatino, V., Marotta, P., Ambrosino, L., Miralto, M. & Ferrante, M. I. De novo transcriptome assembly and gene annotation of a lipoxygenase knock-down mutant in the diatom Pseudo-nitzschia arenysensis. Zenodo https://doi.org/10.5281/zenodo.10026213 (2023).
Lischer, H. E. L. & Shimizu, K. K. Reference-guided de novo assembly approach improves genome reconstruction for related species. BMC Bioinformatics 18, 1–12 (2017).
Joint Genome Institute, 2021. CSP:2021: 100 Diatom Genomes. https://jgi.doe.gov/csp-2021-100-diatom-genomes/.
Karsenti, E. et al. A holistic approach to marine eco-systems biology. PLoS Biol. 9 (2011).
Acknowledgements
PM was partially supported by the Gordon and Betty Moore Foundation through grant no. 7978 to MIF and by the grant PON PRIMA-Rafforzamento Del Capitale Umano, n. CIR01_00029. This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 654008.
Author information
Authors and Affiliations
Contributions
V.S. and M.I.F. conceived the experiments, V.S. produced the data. L.A. and M.M. carried out the bioinformatic analyses. P.M., V.S., L.A. and M.I.F. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Marotta, P., Sabatino, V., Ambrosino, L. et al. De novo transcriptome assembly of a lipoxygenase knock-down strain in the diatom Pseudo-nitzschia arenysensis. Sci Data 11, 522 (2024). https://doi.org/10.1038/s41597-024-03375-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03375-0