Abstract
Branch retinal vein occlusion (BRVO) is the most prevalent retinal vascular disease that constitutes a threat to vision due to increased venous pressure caused by venous effluent in the space, leading to impaired visual function. Optical Coherence Tomography Angiography (OCTA) is an innovative non-invasive technique that offers high-resolution three-dimensional structures of retinal blood vessels. Most publicly available datasets are collected from single visits with different patients, encompassing various eye diseases for distinct tasks and areas. Moreover, due to the intricate nature of eye structure, professional labeling not only relies on the expertise of doctors but also demands considerable time and effort. Therefore, we have developed a BRVO-focused dataset named Soul (Source of ocular vascular) and propose a human machine collaborative annotation framework (HMCAF) using scrambled retinal blood vessels data. Soul is categorized into 6 subsets based on injection frequency and follow-up duration. The dataset comprises original images, corresponding blood vessel labels, and clinical text information sheets which can be effectively utilized when combined with machine learning.
Similar content being viewed by others
Background & Summary
Branch retinal vein occlusion is recognized as one of the most prevalent sight-threatening retinal vascular diseases observed in adult cohort1,2,3. The reported prevalence of BRVO is estimated to be 0.4% worldwide with no gender difference3,4. As the growth of the age increased risk, and patients with monocular BRVO have a 10% risk of developing retinal vein occlusion (RVO) in the contralateral eye within a three-year period5. The pathogenesis of BRVO is multifactorial and has not been fully determined6. BRVO can lead to macular edema, insufficient blood supply to the retina, and thus blurred or impaired vision7. In some cases, BRVO may cause retinal oxygen deprivation, which stimulates the formation of potentially new blood vessels8.Theses vessels are fragile and prone to rupturing and bleeding, further damaging vision9,10. BRVO may also lead to complications such as glaucoma, and vitreous hemorrhage. An example of the different imaging modes is shown in Fig. 111. Therefore, although it may be asymptomatic in the early stages, can have serious effects on vision if left untreated12. The clinical treatment of BRVO patients involves two practical aspects: firstly, reducing macular edema and improving visual acuity; secondly, minimizing the occurrence of complications13. Currently, there are three anti-VEGF drugs available: ConberCept, Lucentis, and Eylea. Large clinical studies have demonstrated that all three drugs yield positive outcomes, with over 50% of patients experiencing significant vision improvement14,15.
Fluorescein Angiography (FA) helps characterize the retinal vascular system, including degree of non-perfusion, macular ischemia, macular edema, and leakage16. Although FA currently serves as the gold standard method for clinical evaluation of retinal perfusion, problems such as internal retinal bleeding, invasive nature and time-consuming inability to be repeated at each visit, as well as its incapability to visualize retinal capillary plexuses at different depths and the potential adverse effects associated with intravenous dye injection can make it difficult to obtain high-quality FA8,17,18. In contrast, OCTA is a recently developed non-invasive technique that utilizes a split-spectrum amplitude decorrelation vascular imaging algorithm16,19. It provides both structural and vascular information, enabling imaging of the superficial layer, deep layer, outer Retina layer, and choroidalcapillary layer of vessels, respectively20. Several studies investigating OCTA in BRVO patients have reported microvascular abnormalities such as telangiectasias, microaneurysms, collateral vessels, and nonperfusion zones20,21. Furthermore, these studies have demonstrated that OCTA can depict abnormal vascular changes equally or even more effectively than FA21,22,23.
Given the recent advancements in artificial intelligence (AI) technology in computer-aided disease diagnosis and treatment, multiple significant advancements started to focus on applying AI methods for automated analysis of BRVO. To foster the development of OCTA image analysis and processing technology, Nanjing University Of Science And Technology and Cixi Institute of Biomedical Engineering has publicly released OCTA-50024,25 and Rose26, respectively. However, these datasets were collected from a single visit to various patients and encompassed a wide range of eye diseases. Furthermore, the creation of these labels heavily relied on the manual efforts of domain experts, which consumed significant amounts of time, energy, and financial resources. To the best of our knowledge, prior to this study, there was no publicly available OCTA database specifically focused on a particular ophthalmic disease with multiple diagnostic capabilities. In order to foster research on BRVO disease, we have released Soul dataset comprising image-text data from 53 patients. This study was received approval from the Institutional Review Board at the Affiliated hospital of Shandong Second Medical University (wyfy-2020-ky-11), following the principles outlined in the Declaration of Helsinki and with the include informed consent of all participants.
Finally, our contributions include:
-
(1)
The datasets we constructed consist solely of patients with BRVO, all of whom have been diagnosed with BRVO disease due to vascular loss or macular edema, resulting in various changes in image data. These datasets do not include normal disease-free eye imaging data.
-
(2)
Multiple pre-operative and post-operative follow-up visits were performed and the data were divided into different subsets. This allows for longitudinal analysis of patient changes and expands beyond a single segmentation task. Researchers can now select appropriate data for clinical research purposes such as disease diagnosis and prognosis after completing the segmentation task.
-
(3)
We have developed a comprehensive framework for automatic vascular label formation that utilizes model integration technology and deep learning knowledge to generate high-quality vascular labels. In comparison to alternative approaches, this framework not only attains a high level of accuracy but also significantly mitigates the expenses associated with manual annotation.
Methods
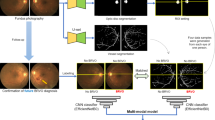
We categorize the generation of Soul datasets into three primary stages: initial clinical data acquisition, subsequent data filtering and pre-processing, and ultimately the implementation of a Human machine collaborative annotation framework (HMCAF). The overall workflow is illustrated in Fig. 2.
Clinical data acquisition
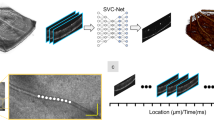
Soul is a multi-diagnosis dataset collected from 53 subjects. The data collected at baseline include OCTA retinal images among BRVO patients in ages between 31 and 82 years old retrospectively collected from the Affiliated hospital of Shandong Second Medical University between 2020 to 2021. This study was received approval from the Institutional Review Board at the Affiliated hospital of Shandong Second Medical University (wyfy-2020-ky-11), following the principles outlined in the Declaration of Helsinki and with the include informed consent of all participants. OCTA images were included if (1) They do not have noticeable quality issues, such as severe smudges, artifacts, out-of-focus, blurriness, incorrect exposure, etc., that would affect the clarity of the observed target area. (2) To ensure spatial alignment of longitudinal tracking data, the macular fovea area is first labeled by a professional ophthalmologist during each patient visit. Subsequently, the 6*6mm area of the fovea of the macula is identified by automatic spatial alignment using the Optovue Angio OCT RTVueXR, a specialized ophthalmic device. This process uses high-resolution optical coherence tomography angiography to ensure accurate alignment of data from each image, helping doctors better monitor the progression of lesions and develop more effective treatment plans. An example is shown in Fig. 3, The results of multiple imaging data comparison showed that the blood vessels were reconstructed to a certain extent after multiple treatments. (3) Since the anti-VEGF Ranibizumab Injections drug is only valid for one month, in order to ensure the validity of follow-up data, only the information of visits on the day of Injections and visits within one week of Injections are kept in the follow-up data, and the follow-up should be ensured within 2-3 days after surgery as far as possible. Images were excluded if they showed any evidence of treatment, severe exposure abnormalities, severe refractive interstitial opacities, large-scale contaminations or if information about its origin was missing.
Text Record
The data primarily encompasses diverse medical information of patients, encompassing collection numbers, gender, age, diseased eye (left and right), disease progression, surgical dates, follow-up visit count, macular center thickness measurements, visual acuity assessments etc., with the corresponding records stored in an Excel file. To ensure participant anonymity and confidentiality, personal identifiers such as names are removed which can identify the subjects identity.
Image
The OCTA images typically acquired and stored grayscale in nature, saved in JPG format. The scanning process employed the Optovue Angio Oct RTVueXR system. It took approximately three years to collect and annotate these images. All subjects have complete registration information, with diagnosed diseases provided by ophthalmologists. Figure 4 (left) illustrates an sample of the scanned images alongside Projection maps at various levels. Although OCTA can produce projection Superificial layer (SVC), Deep layer (DVC), Outer Retina layer, and Choriocapillaris layer according to different retinal projection map, since BRVO disease image features are mostly based on the Superificial layer information, we have only take the SVC layer as the research object and build labels for its images. The retinal hierarchy corresponding to different projections is shown in Table 1 and Fig. 4 (right).
Model Pre-training and Data pre-processing
Soul encompasses three subsets, which have been categorized based on the number of injections and follow-up periods. These subsets include Soul-1(s1t1 & s1t2), Soul-2(s2t1 & s2t2), and Soul-3(s3t1 & s3t2), corresponding to patients who underwent a minimum of one, two, and three surgical treatments respectively. The characteristic of HMCAF is to reduce the time investment of expert manual annotation as much as possible while ensuring the authenticity and accuracy of labels. To ensure image quality and model performance, before original images imported into the baseline models of the HMCAF framework, A series of data processing operations such as data normalization, scale transformation, brightness change and contrast change were carried out. All images were uniformly cropped to eliminate any unused or unimportant boundaries and resized images with 304 * 304 pixels samed as OCTA-500 and ROSE. Before generating initial labels, the baseline model in the HMCAF framework needs to be pre-trained on the OCTA dataset to produce more accurate results. We pre-trained each of the four baseline models on the open source ROSE dataset and achieve optimal performance on the ROSE dataset. The baseline model performance is shown in Table 2.
Human machine collaborative annotation framework
In order to balance labeling accuracy and expert annotation cost, we propose a framework for automatically generating Pseudo-expert label, which consists of two modules: deep learning module and manual correction medules. Its framework is shown in the following Fig. 5.
Machine learning
For the beginner learner, we select the basic framework model OCTA-NET26, U-Net27, AttResU-Net28 and AttU-Net29, which are pre-train on the ROSE-1 dataset using two-level labels of the SVC, to generate primary labels. However, the results are controversial due to the nature of the data for tiny blood vessel. Subsequently, we used the weighted fusion method to integrate the results of different models to obtain an improved fine vessel fusion label, as shown in Fig. 6.
Human correction
Through deep learning, we have achieved labeling results with a certain level of accuracy. However, the inherent characteristics of diseases, such as patients’ poor fixation and other factors. Hence, to further enhance the accuracy and clinical applicability of our results, our ophthalmologists employ Labelme software (https://github.com/labelmeai/labelme)30 to correct the fusion label results (including additions, deletions, modifications, etc.). An example of artifacts and a comparison chart of expert corrections are shown in the Fig. 7.
Data Records
Soul is available on Figshare31. All personal information that could be used to identify the patients was removed before preparation. The dataset details are presented in Table 3, encompassing the mean age of patients, gender distribution proportions, and image counts.
The corresponding diagram of the data subset is shown in the figure below.The dataset comprises projection maps, four types of text labels, and pixel-level labels constructed through human machine collaboration. In Fig. 6 on the left, the subgraph represents three surgeries denoted as s1 (includes s1t1, s1t2), s2 (includes s2t1, s2t2), and s3 (includes s3t1, s3t2) respectively, with a time interval greater than or equal to one month (i.e., s2 - s1 >= 1 month; s3 - s2 >= 1 month). The term “s1t1” refers to the first follow-up after the initial operation, while subsequent terms indicate different surgeries and their respective follow-up visits. Herein, it is ensured that s1 - s1t1 >= 7 days and s1 - s1t2 >= 14 days.
In order to satisfy one operation and two follow-up visits, 36 individuals must be included in a total of 48; similarly, for the subgraph depicted in Fig. 8 on the left, Soul’s structure is illustrated. Additionally, Fig. 9 provides a sample set of data for pre-operative and post-operative follow-up diagnoses with multiple samples.
Technical Validation
Data
The OCTA scans were selected retrospectively by a search of the digital files from a public university hospital and a private imaging practice. For the Optovue Angio OCT device,the examinations were performed from Nov.20.2020 to Dec.30.2021.
Inclusion criteria
OCTA images was performed using the angio retina mode. A 6 × 6mm scan, centered on the fovea, was performed in all study eyes for diagnosing branch retinal vein occlusion. File availability in the Picture Archive and Communication System (PACS) from medical center.
Exclusion criteria
Artifacts that prevented the ophthalmologist from visually interpreting the images. Examinations with fully or partially corrupted files. There was no restriction on age, patient status(inpatients or outpatients) or any other inclusion or exclusion criteria different from those mentioned.
Model selection
We chose the widely recognized U-Net basic model and the classical attention module model as our baseline models, along with the latest retinal vascular segmentation model OCTA, to ensure that our reusable baseline models are not only classical but also incorporate state-of-the-art research features.
Docter announction
The label results of model reuse were manually reviewed and corrected by ophthalmologists specialized in BRVO-related diseases.
Usage Notes
Readers who wish to download the dataset may do so at figshare31. We hope that this published dataset will be available to more researchers and encourage more authors to publish their optimized codes and models, which will contribute to the development and advancement of the field of BRVO disease. Although the dataset we constructed does not have a significant improvement in the number of datasets, our dataset gives multiple follow-up data for the same patient, including image data and corresponding text labels. This improvement allows us to use multiple follow-up data to assist doctors in disease diagnosis or prognosis assessment, and to use image-text multimodal data for related research. The HMCAF we built can greatly reduce manual input while ensuring accuracy. The framework we built is not limited to a certain kind of data, it has a good generalization effect on various clinical data, it is not only suitable for OCTA image data, but also can be well generalized to CT data, MRI data and so on; This process can achieve better results in the relatively precise segmentation task of blood vessels, so the segmentation of lesions such as optic discs and pulmonary nodules can also achieve better results. Clear instructions for acquisition and segmentations are given for readers who wish to increase the size of the dataset.
Code availability
The code is publicly available in the following GitHub repository: https://github.com/CMAIBITU/Soul. The repository encompasses python version, which contains how to preprocess the original image, how to extract the ROI region and pre-trained state-of-the-art deep learning models of OCTA image segmentation on ROSE publicic dateset.
References
Choi, Y.-J., Jee, D. & Kwon, J.-w Characteristics of major and macular branch retinal vein occlusion. Scientific Reports 12, 14103 (2022).
Kang, M. S., Kim, S. Y., Park, S. W., Byon, I. S. & Kwon, H. J. Association between capillary congestion and macular edema recurrence in chronic branch retinal vein occlusion through quantitative analysis of oct angiography. Sci Rep 11, 19886, https://doi.org/10.1038/s41598-021-99429-z (2021).
Laouri, M., Chen, E., Looman, M. & Gallagher, M. The burden of disease of retinal vein occlusion: review of the literature. Eye 25, 981–988 (2011).
Lim, L. L. et al. Prevalence and risk factors of retinal vein occlusion in an asian population. British Journal of Ophthalmology 92, 1316–1319 (2008).
Yau, J., Lee, P., Wong, T., Best, J. & Jenkins, A. Retinal vein occlusion: an approach to diagnosis, systemic risk factors and management. Internal medicine journal 38, 904–910 (2008).
Rehak, J. & Rehak, M. Branch retinal vein occlusion: pathogenesis, visual prognosis, and treatment modalities. Current eye research 33, 111–131 (2008).
Christoffersen, N. L. & Larsen, M. Pathophysiology and hemodynamics of branch retinal vein occlusion. Ophthalmology 106, 2054–2062 (1999).
Gualino, V. et al. Optical coherence tomography, fluorescein angiography, and diagnosis of choroidal neovascularization in age-related macular degeneration. Retina 39, 1664–1671, https://doi.org/10.1097/IAE.0000000000002220 (2019).
Virmani, R. et al. Atherosclerotic plaque progression and vulnerability to rupture: angiogenesis as a source of intraplaque hemorrhage. Arteriosclerosis, thrombosis, and vascular biology 25, 2054–2061 (2005).
Han, H.-C. Twisted blood vessels: symptoms, etiology and biomechanical mechanisms. Journal of vascular research 49, 185–197 (2012).
Jaulim, A., Ahmed, B., Khanam, T. & Chatziralli, I. P. Branch retinal vein occlusion: epidemiology, pathogenesis, risk factors, clinical features, diagnosis, and complications. an update of the literature. Retina 33, 901–910 (2013).
Bressler, N. M. Early detection and treatment of neovascular age-related macular degeneration. The Journal of the American Board of Family Practice 15, 142–152 (2002).
Shahid, H., Hossain, P. & Amoaku, W. The management of retinal vein occlusion: is interventional ophthalmology the way forward? British Journal of Ophthalmology 90, 627–639 (2006).
Lalwani, G. A. et al. A variable-dosing regimen with intravitreal ranibizumab for neovascular age-related macular degeneration: year 2 of the pronto study. American journal of ophthalmology 148, 43–58 (2009).
Thach, A. B., Yau, L., Hoang, C. & Tuomi, L. Time to clinically significant visual acuity gains after ranibizumab treatment for retinal vein occlusion: Bravo and cruise trials. Ophthalmology 121, 1059–1066 (2014).
Chalam, K. & Sambhav, K. Optical coherence tomography angiography in retinal diseases. Journal of ophthalmic & vision research 11, 84 (2016).
Tey, K. Y. et al. Optical coherence tomography angiography in diabetic retinopathy: a review of current applications. Eye and Vision 6, 1–10 (2019).
Raabe, A., Beck, J., Gerlach, R., Zimmermann, M. & Seifert, V. Near-infrared indocyanine green video angiography: a new method for intraoperative assessment of vascular flow. Neurosurgery 52, 132–139 (2003).
De Carlo, T. E., Romano, A., Waheed, N. K. & Duker, J. S. A review of optical coherence tomography angiography (octa). International journal of retina and vitreous 1, 1–15 (2015).
Kashani, A. H. et al. Optical coherence tomography angiography: A comprehensive review of current methods and clinical applications. Progress in Retinal and Eye Research 60, 66–100, https://doi.org/10.1016/j.preteyeres.2017.07.002 (2017).
Tan, A. C. et al. An overview of the clinical applications of optical coherence tomography angiography. Eye 32, 262–286 (2018).
Deng, Y. et al. Quantitative analysis of retinal microvascular changes after conbercept therapy in branch retinal vein occlusion using optical coherence tomography angiography. Ophthalmologica 242, 69–80 (2019).
Borrelli, E., Sarraf, D., Freund, K. B. & Sadda, S. R. Oct angiography and evaluation of the choroid and choroidal vascular disorders. Progress in retinal and eye research 67, 30–55 (2018).
Li, M. et al. Image projection network: 3d to 2d image segmentation in octa images. IEEE Transactions on Medical Imaging 39, 3343–3354 (2020).
Li, M. et al. Ipn-v2 and octa-500: Methodology and dataset for retinal image segmentation. arXiv preprint arXiv:2012.07261 5, 16 (2020).
Ma, Y. et al. Rose: a retinal oct-angiography vessel segmentation dataset and new model. IEEE transactions on medical imaging 40, 928–939 (2020).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18, 234–241 (Springer, 2015).
Lafraxo, S., Souaidi, M., El Ansari, M. & Koutti, L. Semantic segmentation of digestive abnormalities from wce images by using attresu-net architecture. Life 13, 719 (2023).
Wang, S., Li, L. & Zhuang, X. Attu-net: attention u-net for brain tumor segmentation. In International MICCAI Brainlesion Workshop, 302–311 (Springer, 2021).
Russell, B. C., Torralba, A., Murphy, K. P. & Freeman, W. T. Labelme: a database and web-based tool for image annotation. International journal of computer vision 77, 157–173 (2008).
Xue, J., Feng, Z. & Xia, J. Soul: An octa dataset based on a human-machine collaborative annotation framework. Figshare https://doi.org/10.6084/m9.figshare.24893358.v3 (2024).
Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China (2022YFC2403902, and 2023YFC3502604), the National Natural Science Foundation of China (82174533, and 82374302), Key R&D project of Ningxia Autonomous Region (2022BEG02036), Shandong Province Medical and Health Science and Technology Project (2023BJ000045),the Fundamental Research Funds for the Central Universities (2024JBMC007).
Author information
Authors and Affiliations
Contributions
Jingyan Xue, Zhenhua Feng and Shuana Wang collected the data from hospitals, organized related materials, and created data sets. Jingyan Xue and Lili Zeng normalized the clinical data, preprocessed the images and used the machine learning method to generate the initial labels. Zhenhua Feng and Aijun Deng reviewed and corrected the model integration labels. Xuezhong Zhou and Jiana Xia supervised this work and provided important guidance on the writing of the article. Jingyan Xue and Jianan Xia prepared the manuscript, figures and tables. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it.The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xue, J., Feng, Z., Zeng, L. et al. Soul: An OCTA dataset based on Human Machine Collaborative Annotation Framework. Sci Data 11, 838 (2024). https://doi.org/10.1038/s41597-024-03665-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-024-03665-7