Abstract
Progress in the acquisition of massive sets of molecular data and in the bioinformatic capabilities for their processing have revolutionised species identification, filling gaps in crucial areas such as taxonomy, phylogenetic inference, biogeography, and even biodiversity conservation. Advanced DNA sequencing and metabarcoding have uncovered previously hidden diversity, although their effectiveness is highly dependent on the accuracy of reference DNA databases at local and regional scales. The compilation of information on freshwater fishes from the Magdalena River basin is an important milestone in improving our knowledge of the genetic and taxonomic diversity of a highly endemic region in the Neotropical context. Here, we share DNA data from 1,270 specimens representing 183 species, cross-referenced with complete collecting and catalogue information, along with high resolution photographs of voucher specimens when alive. This collection of multiple sources of information based on fish specimen records not only contributes to future research on Neotropical fish systematics and ecology, but also to conservation decisions in one of the South American rivers with a highest level of endemism.
Similar content being viewed by others
Background & Summary
The landscape of the northern Andes in Colombia is shaped by three mountain ranges (Western, Central, and Eastern), forming two extensive inter-Andean valleys drained by the Magdalena and Cauca rivers1. These valleys collectively constitute the Magdalena River basin (MRB), which is the largest trans-Andean basin. These ranges played a crucial role in determining the evolution of a unique species assemblage, constituting the ichthyofauna with the highest proportion of endemic species in Colombia2,3. This region encompasses a diverse array of aquatic environments along latitudinal and altitudinal gradients, forming a mosaic of habitats for a wide diversity of fish species, some of which are particularly significant in local fisheries4,5,6. The fish species in this region display a broad repertoire of adaptations to cope with vastly contrasting habitat conditions7. These adaptations are frequently found in specialised species, narrowly distributed in very specific habitats, unique to this mountainous region, where the level of endemism is notably high8. However, our understanding of the diversity and distribution of fish species does not progress as rapidly as the anthropogenic transformations occurring in these aquatic environments9. Therefore, developing data tools that enhance the taxonomic identification of fish species is essential to guide conservation decisions in the MRB. The lack of such information could result in the exclusion of considerations centred on the fish fauna, in development projects, with potentially severe consequences10.
Scientific research on fishes from the MRB was initiated by Humboldt (1805), who provided taxonomic descriptions of several endemic species. Taxonomic information was subsequently generated by foreign ichthyologists11, mostly naturalists from museums established in large European cities12,13,14,15,16,17. Notably, the contributions of the German-American ichthyologist Carl H. Eigenmann were based on an ambitious ichthyological reconnaissance of the territory that stands out as a landmark in the study of Colombian fishes18,19,20,21,22. Recent studies conducted by local researchers are expanding the taxonomic knowledge of the fish species from the Colombian Andes23,24,25. Historically, fish species have been identified using morphological traits. However, challenges arise when dealing with cryptic or morphologically similar species, particularly within genera of complex taxonomy such as Astroblepus, Astyanax, Characidium, Chaetostoma, Hemibrycon, Trichomycterus, among others26,27. Given these challenges, identifications based solely on morphology have required remarkable expertise in specific taxa to minimise the risk of identification errors. Therefore, new technologies and diagnostic tools such as digital image processing and pattern recognition techniques, characterization of acoustic or electric organ signals, and molecular methods are necessary28,29,30.
In recent decades, molecular methods have played a pivotal role in the discovery of hidden diversity, as identification becomes independent of the taxonomic expertise of the researcher31 or the life stage of the species32. These advancements have facilitated the integration of diverse fields, spanning taxonomy, conservation, invasive species detection, and biogeography33,34,35. Notably, the widespread application of advanced DNA sequencing technologies has allowed detailed analyses of genes and complete genomic sequences36,37,38,39. One significant breakthrough is the adoption of the metabarcoding approach, enabling the simultaneous identification of multiple samples40,41. This approach has extended its applications to studying entire ecological communities through environmental DNA sampling42,43,44. However, the efficacy of these procedures hinges on the availability and accuracy of DNA reference databases, some of which are public and crucial for bioinformatic applications (i.e. Barcode of Life BOLD: https://www.boldsystems.org/, GenBank: https://www.ncbi.nlm.nih.gov). The utilisation of these reference databases has witnessed a substantial increase worldwide in recent years45,46,47. These databases are linked to specimens catalogued in natural history museums, allowing reexamination to verify or revise taxonomic identifications. For fishes in the MRB, there are existing public DNA databases of the mitochondrial Cytochrome c Oxidase subunit I (COX1)48, both in the BOLD and GenBank platforms26,27,31, as well as complete mitochondrial genomes49. These resources contribute significantly to the understanding of the fish species diversity and enhance the accuracy and reliability of species identification.
The Magdalena biodiversity is mostly reflected and preserved in the country’s valuable biological collections. Despite the incomplete taxonomic representation of the fish diversity in biological collections, in part due to the continuous description of new taxa, cases such as the Collection of Ichthyology of the University of Antioquia (CIUA) stands out for its remarkable taxonomic and geographical representation of MRB fishes (208 species and 815 sites), being crucial for precise taxonomic identification, effective conservation, and management (Fig. S1). Valuing Colombia’s natural and cultural heritage, the CIUA’s extensive collection not only serves as a repository for biodiversity documentation, but also plays a role in advancing scientific research, education, and public outreach initiatives related to MRB fishes. To disentangle the evolution and taxonomy of complex fish groups as those already referred before, the University of Antioquia and the hydropower company Empresas Públicas de Medellín (EPM) began a collaboration in 2017 to establish a genetic reference database of the fishes preserved in the CIUA collection. This database has been built from specimens collected during environmental monitoring in the areas of influence of EPM’s hydropower plants and in expeditions intended to search representative topotypes of endemic species.
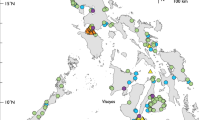
Our genetic database was built with specimens from 297 localities, encompassing a wide array of aquatic environments from the Atrato, Catatumbo, Cauca, Dagua, and a significant representation from the MRB (Fig. 1). The sampling conducted in these basins holds fundamental significance in comparing the taxonomic identification in use within the MRB. A total of 1,270 specimens were taxonomically classified, and preserved according to the procedures described in the methods section. Tissue samples were collected, and their DNA was extracted and sequenced for three mitochondrial regions: COX1, 12S ribosomal RNA, and 16S ribosomal RNA large subunit (Table 1). The specimens were catalogued with extensive metadata (Supplementary Table 1), resulting in a DNA database representing at least 183 species, 90 genera, and 36 families of fishes (Fig. 2). Additionally, the collection includes photographs corresponding to 168 species (Fig. 3). Species identifications were rigorously verified, based on phenotypic and genotypic data (see Methods and Technical Validation sections).
The data obtained have already led to the publication of taxonomic descriptions of new species50. Topotypes, accounting for 39% of the species in our dataset (Fig. 3), were crucial to ensure a reliable source of taxonomic validation. The dataset includes DNA sequences from 54 species not previously available in GenBank or BOLD for any loci, along with additional species representing the first publicly available COX1, 12S, and 16S sequences for that taxon (Table 1). Moreover, we have initiated the construction of a dataset of mitochondrial genomes with results for 10 endemic fish species, some of which are important for fisheries or are currently classified as endangered species (Fig. 4). This comprehensive dataset contributes to advancing our understanding of the genetic diversity of fish species in the region. The 27.88% of fish species in this dataset have been classified according to their conservation threat status (51 species), while 66.67% are labelled as data deficient (122 species), and the remaining 5.46% have not been evaluated yet (10 species) (Fig. 5). The list of species and their IUCN status is available in Supplementary Table 1. These species are locally endemic, with low abundance (estimated by their capture frequency), or correspond to recently described species, or species in undersampled aquatic environments. Consequently, the lack of information on distribution, ecology, population trends, and threats, hinders conservation strategies. However, it is hoped that this database will contribute to providing the necessary tools for more informed assessments within the International Union for Conservation of Nature (IUCN; https://www.iucnredlist.org/), and granting legal status for conservation policies. In addition, 11 species of introduced fishes (non-native) that have been identified as invasive are incorporated into the database (details on these species can be found in Supplementary Table 1). This information is essential for the recognition and monitoring of invasive species by DNA metabarcoding, a technique that facilitates early detection and large-scale monitoring of these habitats.
This comprehensive database encompasses approximately 65% (153 species) of the estimated fish diversity in the MRB51. CIUA is actively contributing to the expansion of this coverage by publishing new DNA sequences on public platforms, a project outlined in detail on GenBank, and CavFish (https://cavfish.unibague.edu.co/) (details on these species can be found in Supplementary Table 1). This ongoing effort aims to continually increase the representation of fish species in the region, enhancing the utility and completeness of the database. This collection of information serves as an indispensable reference for the knowledge of Colombian fishes, providing valuable resources for a diverse community of users with varied interests. This resource not only contributes to ongoing research but also lays the groundwork for future investigations. Moreover, it will bring key elements for taxonomic research to unhide undescribed species in multiple lineages, completed with comprehensive collection data and detailed photographic catalogues of the voucher specimens of the genetic sequences constituting the database. Simultaneously, this database plays a crucial role in refining the precision of results in the growing number of studies employing DNA metabarcoding. As molecular methods become increasingly prevalent in ecological studies, the comprehensive and accurate data provided by our collection of information will undoubtedly contribute to more robust and reliable outcomes in research focusing on the intricate dynamics of the fish species from one of the most biodiverse regions in the world.
Methods
This study was conducted with the recommendations and approval of the Ethics Committee on Animal Testing of the University of Antioquia (CEEA). The protocol was reviewed and approved on November 14, 2017 by CEEA, and updated on February 9, 2021. Specimen collection was endorsed by the Ministry of Environment of Colombia through the Non-Commercial Scientific Research Permit granted to University of Antioquia (Resolution 0524 of May 27, 2014).
Sampling design and specimen collection
The database includes captures of fishes from the year 2010 through 2023 (Supplementary Table 1). These captures were in diverse aquatic ecosystems distributed between 0 and 3,500 metres above the sea level (Supplementary Table 1), including rivers, streams, creeks, floodplain lakes, and Andean reservoirs. Geographic coordinates were recorded at each sampling location using a satellite geopositioner (GPS), calibrated to the WGS84 datum. Due to the selectivity of the catch method for different fish species and body size, we standardise the catch effort for each environment (flowing channels, floodplains lakes, and reservoirs). We used the same catch effort for each environment. In flowing channels (creeks, streams, and rivers), the catch effort was 30 sets with each of the three cast nets (with different mesh sizes: 0.5, 1.5, and 3.5 cm), plus 60 min of sweeps with portable electric fishing equipment of pulsating DC current (340 V, 1–2 A) along 100 m of the flowing channel. In the littoral zones of floodplains, lakes, and reservoirs, the sampling was made by deploying two gill nets, each measuring 100 m long and 3 m high, during six-hour periods when possible. This time was adjusted by constraints due to access and security regulations in the reservoirs, or the maximum time allowed by local communities in floodplains lakes. Each gill net featured ten different mesh sizes (1–10 cm between opposite knots) to improve the chance of capturing a wide spectrum of fish species and body sizes.
Captured specimens were immersed in a light anaesthesia bath using eugenol (12.5 mg/l), in a stock solution of 1:9 (eugenol:ethanol), to reduce stress and mortality associated with handling52. Subsequently, at least one specimen per species at each locality was selected for live photographic documentation using the Photafish System53. Photographs were taken by J. L. Londoño-López, J. E. García-Melo, and J. G. Ospina-Pabón using SONY Alpha 6000, SONY Alpha 7III, SONY Alpha 7RIV, and Nikon d5500 camera bodies, with 90 mm and 60 mm macro lenses, under flash lighting. The specimens were then sacrificed receiving double dosage of eugenol, until they lost swimming activity and respiration. Tissue samples from each specimen were extracted and preserved at 96% ethanol, and whole voucher specimens were fixed with 10% formalin. Voucher specimens were subsequently transferred to 75% ethanol for long-term preservation and catalogued at CIUA. Associated tissues and DNA extractions were stored at −82 °C and −20 °C, respectively, in CIUA biorepository freezers.
Taxonomic identification and validation
Field taxonomic identifications were performed on fresh specimens and then confirmed in the laboratory using the same preserved specimens. This process involved regional or taxon-specific keys complemented by systematic and taxonomic reviews at the family or genus level (when available), as well as original species descriptions and redescriptions. For some taxonomic groups, these taxonomic identifications were validated by specialists: H. D. Agudelo-Zamora (Characidium), J. G. Albornoz-Garzón (Stevardiinae), T. P. Carvalho (Bunocephalus), M. C. Castellanos-Mejía (Rineloricaria), C. DoNascimiento (Heptapteridae, Pimelodidae, Trichomycteridae), C. A. García-Alzate (Hyphessobrycon), M. A. Hernández-Cortés (Heptapteridae), F. C. T. Lima (Bryconidae), N. K. Lujan (Hypostominae), V. M. Medina-Ríos (Trichomycterus), A. Méndez-López (Bryconidae), J. G. Ospina-Pabón (Astroblepidae), A. T. Thomaz (Argopleura). Ordinal classification and valid names adhere to Near and Thacker54 and DoNascimiento et al.51, respectively.
Taxonomic identification of voucher specimens was confirmed by comparing newly generated sequences with those available in public repositories (GenBank), as detailed in the Technical Validation section below. Genomic DNA was extracted from tissue samples using the DNeasy Blood and Tissue kit (Qiagen) and the GeneJET Genomic DNA Purification kit (Thermo Scientific), following the manufacturers’ protocols. DNA samples were amplified by PCR (Table 2). PCR reactions were conducted in a 30 µl reaction volume, consisting of 0.6 µl of each primer (10 mM), 0.6 µl of dNTPs (10 mM), 3 µl of reaction buffer (10x), 0.3 µl of OneTaq® DNA polymerase (5U/µl), and 3 µl of genomic DNA. The concentration of MgCl2 varied according to the molecular marker (Table 2). The PCR temperature cycle consisted of an initial 5 min step at 95 °C, followed by 35 cycles of 45 s at 94 °C, 1 min for primer hybridization (temperatures specified in Table 2), 1 min at 72 °C, and a final extension phase of 10 min at 72 °C.
Data Records
The MRB fish database encompasses a comprehensive set of information resources, including; (1) preserved specimens catalogued and deposited in the CIUA55, (2) DNA and tissue banks obtained from voucher CIUA catalogued specimens (details on these species can be found in Supplementary Table 1), (3) a full-resolution photographic collection of voucher CIUA catalogued specimens photographed when still alive, from which a selection of representative photographs (at lower image resolution for optimal web-page visualisation) of each species from each collecting site (usually corresponding also to voucher specimens of published genetic sequences) is available at the CavFish project website (https://cavfish.unibague.edu.co/) (details on these species can be found in Supplementary Table 1), and (4) an edited database of genetic sequences corresponding to three mitochondrial loci (COX1, 12S, and 16S) along with complete mitochondrial genomes publicly available in GenBank56. These information resources ensure wide and easy accessibility for research and educational purposes, promoting an open-data approach for scientific and conservation collaborations, and further exploration and knowledge construction of the ichthyofauna diversity from Colombia and the Neotropics.
Technical Validation
Consensus sequences were assembled and edited using Geneious Prime v. 2023.1.2 (http://www.geneious.com) and aligned using the G-INS-i plugin in MAFFT v.7, with default parameters57. Subsequently, Maximum Likelihood (ML) phylogenies were individually inferred for each marker in the IQTREE 2 program58. Phylogenetic results were scrutinised to validate sequence identity, primarily based on adherence to well-corroborated monophyletic groups or anomalous phylogenetic placement of individual sequences. In instances where sequences of a single taxon are grouped together, additional validation was conducted using BLAST to ensure high correspondence (>98%) with the taxonomy inferred from morphological traits. For sequences clustered with unexpected lineages, corresponding vouchers were reexamined to confirm their taxonomic identification. These sequences were then cross-checked with BLAST to verify their actual identification. While BLAST facilitated confirmation of the genus, species identification primarily relied on morphological characteristics. This validation process aimed to ensure data accuracy and rectify any potential errors in specimen cataloguing prior to sequencing. It is important to highlight that some sequences matched at the genus level but not at the species level due to the absence of previously deposited sequences in genetic repositories for certain species (e.g., Astroblepus, Hemibrycon, Chaetostoma). This lack of representation underscores the significance of our work, emphasizing the need for further exploration and documentation of these taxa in genetic databases. Sequences falling into this category are classified as either “newly sequenced” or “newly sequenced for each molecular marker”, depending on whether they have been sequenced for one or more loci previously (Table 1).
Mitochondrial genomes were sequenced using the Illumina platform (Illumina Inc., San Diego, CA, USA). The DNA library preparation and sequencing were conducted by Macrogen Company, South Korea (https://dna.macrogen.com). Sequencing libraries were prepared using the TrueSeq Nano 350 bp DNA kit, following manufacturer’s protocol. Raw reads underwent quality filtering using Cutadapt v3.5.859, while medium-depth analysis and detection of alternative alleles were carried out using Bowtie2 v2.4.460 and SAMtools v1.1461. Filtered genomic reads were utilised for de novo assembly of the mitochondrial genome using SPAdes v3.15.362. Graphical annotation and analysis were performed using Mitofish v3.8563 and Proksee64.
Usage Notes
The MRB fish database, an open-access electronic resource, compiles data on cryptic species, particularly those within genera of intricate taxonomy. This database facilitates the review of complex taxa facing taxonomic uncertainties. In addition, it incorporates records of 210 specimens from 18 genera, including Astroblepus, Astyanax, Chaetostoma, Characidium, Creagrutus, Cynodonichthys, Eigenmannia, Hemibrycon, Hypostomus, Knodus, Lasiancistrus, Lycengraulis, Parodon, Poecilia, Pseudopimelodus, Rineloricaria, Sturisomatichthys, and Trichomycterus. However, due to limitations in the taxonomic information (phenotypic and genotypic) currently available, the taxonomic status of these specimens remains unresolved (Supplementary Table 1). As our understanding of fish taxonomy for the MRB increases, additional sequences from these collections will be verified and incorporated into the GenBank project.
We have made an important correction concerning sequences of primers L1484165 and H1591566, used to amplify Cytb. For a long time, these primers names have been incorrectly used to name other primers that amplify 12S gen26,67,68. The precise designation of this sequence is L941-PHE and H2010-VAL (see Table 2), as referenced by Sato69. This clarification is essential to interpret and apply our sequencing results properly.
Code availability
No custom code was used.
References
Albert, J.S. & Reis, R. E. Historical Biogeography of Freshwater Fishes. Introd. to Neotrop. Freshwaters. 20 (2011).
Rodríguez-Olarte, D., Mojica-Corzo, J. I. & Taphorn, D. B. Northern South America: Magdalena and Maracaibo basins. Hist. Biogeogr. Neotrop. Freshw. Fishes 243–258 (2011).
García-Alzate, C., DoNascimiento, C., Villa-Navarro, F. A., García-Melo, J. E. & Herrera-R, G. A. Diversidad de peces de la cuenca del Río Magdalena, Colombia. in Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. (eds. Jiménez-segura, L. F. & Lasso-Alcalá, C. A.) 85–113 (Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, 2020).
Jiménez-Segura, L. F. et al. Freshwater fish faunas, habitats and conservation challenges in the Caribbean river basins of north-western South America. J. Fish Biol. 89, 65–101 (2016).
Herrera-Pérez, J., Parra, J. L., Restrepo-Santamaría, D. & Jiménez-Segura, L. F. The influence of abiotic environment and connectivity on the distribution of diversity in an Andean fish fluvial network. Front. Environ. Sci. 7 (2019).
Valencia-Rodríguez, D. et al. Distribution of diversity of fishes in an Andean fluvial network. Rev. Biol. Trop. 71 (2023).
Conde-Saldaña, C. C., Albornoz-Garzón, J. G., López-Delgado, E. O. & Villa-Navarro, F. A. Ecomorphological relationships of fish assemblages in a trans-Andean drainage, Upper Magdalena River Basin, Colombia. Neotrop. Ichthyol. 15 (2017).
Anderson, E. P. & Maldonado-Ocampo, J. A. A regional perspective on the diversity and conservation of tropical Andean fishes. Conserv. Biol. 25, 30–39 (2011).
Restrepo-Santamaría, D. et al. Bio Anorí, the biological expedition that documented fish diversity after the post-conflict in Antioquia, Colombia. Glob. Ecol. Conserv. 43, e02445 (2023).
Tognelli, M. F. et al. Assessing conservation priorities of endemic freshwater fishes in the Tropical Andes region. Aquat. Conserv. Mar. Freshw. Ecosyst. 29, 1123–1132 (2019).
Posada, A. Los peces. In Estudios científicos del doctor Andrés Posada con algunos otros escritos suyos sobre diversos temas 285–322 (Imprenta oficial, 1909).
Boulenger, G. A. XLIII.—On new siluroid fishes from the Andes of Columbia. Ann. Mag. Nat. Hist. 19, 348–350 (1887).
Boulenger, G. A. XXII.—Descriptions of three new characinid fishes from South-western Colombia. Ann. Mag. Nat. Hist. 7, 212–213 (1911).
Regan, C. T. LVI.—The fishes of the San Juan River, Colombia. Ann. Mag. Nat. Hist. 12, 462–473 (1913).
Steindachner, F. Zur Fischfauna des Magdalenen-Stromes. Anzeiger der Akad. der Wissenschaften Wien 15, 88–91 (1878).
Steindachner, F. Zur Fisch-Fauna des Magdalenen-Stromes. Denkschriften der Kais. Akad. der Wissenschaften Wien, Math. Cl. 39, 19–78 (1879).
Steindachner, F. Zur Fisch-Fauna des Cauca und der Flüsse bei Guayaquil. Denkschriften der Kais. Akad. der Wissenschaften Wien, Math. Cl. 42, 55–104 (1880).
Eigenmann, C. H. Some results from an ichthyological reconnaissance of Colombia, South America. Part II. Indiana Univ. Stud. 18, 1–32 (1913).
Eigenmann, C. H. Eighteen new species of fishes from northwestern South America. Proc. Am. Philos. Soc. 56, 673–689 (1918).
Eigenmann, C. H. The fishes of western South America, Part I. The fresh-water fishes of northwestern South America, including Colombia, Panama, and the Pacific slopes of Ecuador and Peru, together with an appendix upon the fishes of the Rio Meta in Colombia. Mem. Carnegie Museum 9, 1–346 (1922).
Eigenmann, C. & Henn, A. On new species of fishes from Colombia, Ecuador, and Brazil. Indiana Univ. Stud. 24, 231–234 (1914).
Eigenmann, C. H., Henn, A. & Wilson, C. New fishes from western Colombia, Ecuador, and Peru. Indiana Univ. Stud. 19, 1–15 (1914).
Agudelo-Zamora, H. D., Ortega-Lara, A. & Taphorn, D. C. B. Characidium chancoense, a new species of South American darter from the Río Cauca drainage, Colombia (Characiformes: Crenuchidae). Zootaxa 4768 (2020).
Herrera-Collazos, E. E., Galindo-Cuervo, A. M., Maldonado-Ocampo, J. A. & Rincón-Sandoval, M. Three new species of the Eigenmannia trilineata species group (Gymnotiformes: Sternopygidae) from northwestern South America. Neotrop. Ichthyol. 18 (2020).
Londoño-Burbano, A. & Reis, R. E. A taxonomic revision of Sturisomatichthys Isbrücker and Nijssen, 1979 (Loricariidae: Loricariinae), with descriptions of three new species. Copeia 107, 764 (2019).
Ochoa, L. E. et al. Species delimitation reveals an underestimated diversity of Andean catfishes of the family Astroblepidae (Teleostei: Siluriformes). Neotrop. Ichthyol. 18 (2020).
García-Melo, J. E. et al. Species delimitation of neotropical Characins (Stevardiinae): Implications for taxonomy of complex groups. PLoS One 14, e0216786 (2019).
Hernández-Serna, A. & Jiménez-Segura, L. F. Automatic identification of species with neural networks. PeerJ 2, e563 (2014).
Muñoz-Duque, S., López-Casas, S., Rivera-Gutiérrez, H. & Jiménez-Segura, L. Bioacoustic characterization of mating calls of a freshwater fish (Prochilodus magdalenae) for passive acoustic monitoring. Biota Colomb. 22 (2021).
Díaz, J. et al. First DNA barcode reference library for the identification of South American freshwater fish from the lower Paraná River. PLoS One 11, e0157419 (2016).
Restrepo-Gómez, A. M., Rangel-Medrano, J. D., Márquez, E. J. & Ortega-Lara, A. Two new species of Pseudopimelodus Bleeker, 1858 (Siluriformes: Pseudopimelodidae) from the Magdalena Basin, Colombia. PeerJ 8, e9723 (2020).
Rivera-Coley, K., Augusto Reynalte-Tataje, D., Atencio-García, V., Campo, O. & Jímenez-Segura, L. ¿Where do migratory fish spawn in a Neotropical Andean basin regulated by dams? PLoS One 18, e0291413 (2023).
Dufresnes, C., Poyarkov, N. & Jablonski, D. Acknowledging more biodiversity without more species. Proc. Natl. Acad. Sci. 120, e2302424120 (2023).
Van Nynatten, A., Gallage, K. S., Lujan, N. K., Mandrak, N. E. & Lovejoy, N. R. Ichthyoplankton metabarcoding: An efficient tool for early detection of invasive species establishment. Mol. Ecol. Resour. 23, 1319–1333 (2023).
Vences, M., Miralles, A. & Dufresnes, C. Next-generation species delimitation and taxonomy: Implications for biogeography. J. Biogeogr. n/a (2024).
Rangel-Medrano, J. D., Alzate, J. F. & Márquez, E. J. Complete mitochondrial genome of the Neotropical catfish Pseudoplatystoma magdaleniatum (Siluriformes, Pimelodidae). Mitochondrial DNA Part A 27, 4033–4034 (2016).
Landínez-García, R. M., Alzate, J. F. & Márquez, E. J. Complete mitogenome of the Neotropical fish Brycon henni, Eigenmann 1913 (Characiformes, Bryconidae). Mitochondrial DNA Part A 27, 2259–2260 (2016).
Restrepo-Escobar, N., Alzate, J. F. & Márquez, E. J. Mitochondrial genome of the Neotropical catfish Ageneiosus pardalis, Lütken 1874 (Siluriformes, Auchenipteridae). Mitochondrial DNA Part A 27, 2176–2177 (2016).
Yepes-Blandón, J. A. et al. Draft genome assembly for the colombian freshwater bocachico fish, Prochilodus magdalenae. Front. Genet. 13 (2023).
Lozano Mojica, J. D. & Caballero, S. Applications of eDNA metabarcoding for vertebrate diversity studies in Northern Colombian water bodies. Front. Ecol. Evol. 8 (2021).
Polanco, F., A. et al. Detecting aquatic and terrestrial biodiversity in a tropical estuary using environmental DNA. Biotropica 53, 1606–1619 (2021).
Erős, T. et al. eDNA metabarcoding reveals the role of habitat specialization and spatial and environmental variability in shaping diversity patterns of fish metacommunities. PLoS One 19, e0296310 (2024).
Zong, S. et al. Combining environmental DNA with remote sensing variables to map fish species distributions along a large river. Remote Sens. Ecol. Conserv. n/a (2023).
Polanco, F., A. et al. Comparing the performance of 12S mitochondrial primers for fish environmental DNA across ecosystems. Environ. DNA 3, 1113–1127 (2021).
Delrieu-Trottin, E. et al. A DNA barcode reference library of French Polynesian shore fishes. Sci. Data 6, 114 (2019).
Janzen, F. H., Crampton, W. G. R. & Lovejoy, N. R. A new taxonomist-curated reference library of DNA barcodes for Neotropical electric fish (Teleostei: Gymnotiformes). Zool. J. Linn. Soc. 196, 1718–1742 (2022).
Bemis, K. E. et al. Biodiversity of Philippine marine fishes: A DNA barcode reference library based on voucher specimens. Sci. Data 10, 411 (2023).
Mejía-Estrada, M., Jiménez-Segura, L. F., Hernández-Zapata, M. & Soto Calderón, I. D. Contribution to a reference library of DNA barcodes of Colombian freshwater fishes. Biodivers. Data J. 10, e65981 (6AD).
Márquez, E. J., Restrepo-Escobar, N., Yepes-Acevedo, A. J. & Narváez, J. C. Diversidad y estructura genética de los peces de la cuenca del Magdalena, Colombia. in Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible (eds. Jiménez-Segura, L. F. & Lasso, C. A.) 115–157 (Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, 2020).
Castellanos-Mejía, M. C., Londoño-Burbano, A., Ochoa, L. E., García-Alzate, C. A. & DoNascimiento, C. Two new species of Rineloricaria (Siluriformes: Loricariidae) from trans-Andean rivers of Colombia, unveiled through iterative taxonomy. Ichthyol. Herpetol. 112, 429–443 (2024).
DoNascimiento, C. et al. Lista de especies de peces de agua dulce de Colombia/Checklist of the freshwater fishes of Colombia. v2.16. https://doi.org/10.15472/numrso (2024).
Javahery, S., Nekoubin, H. & Moradlu, A. H. Effect of anaesthesia with clove oil in fish (review). Fish Physiol. Biochem. 38, 1545–1552 (2012).
García-Melo, J. E. et al. Photafish system: An affordable device for fish photography in the wild. Zootaxa 4554 (2019).
Near, T. & Thacker, C. Phylogenetic classification of living and fossil ray-finned fishes (Actinopterygii). Bull. Peabody Museum Nat. Hist. 65 (2023).
Jiménez Segura, L. F., Ospina Pabón, J. & DoNascimiento, C. Colección de Ictiología de la Universidad de Antioquia. v5.12. Sib-Colombia https://doi.org/10.15472/lkcff8 (2024).
NCBI GenBank https://identifiers.org/ncbi/bioproject:PRJNA1040268 (2024).
Katoh, K. & Standley, D. M. MAFFT Multiple sequence alignment software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Minh, B. Q. et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Bankevich, A. et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Iwasaki, W. et al. MitoFish and MitoAnnotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 30, 2531–2540 (2013).
Grant, J. R. et al. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51, W484–W492 (2023).
Kocher, T. D. et al. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. 86, 6196–6200 (1989).
Irwin, D. M., Kocher, T. D. & Wilson, A. C. Evolution of the cytochromeb gene of mammals. J. Mol. Evol. 32, 128–144 (1991).
Oliveira, C. et al. Phylogenetic relationships within the speciose family Characidae (Teleostei: Ostariophysi: Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol. Biol. 11, 275 (2011).
Brito, P. S. et al. Cryptic speciation in populations of the genus Aphyocharax (Characiformes: Characidae) from eastern Amazon coastal river drainages and surroundings revealed by single locus species delimitation methods. Neotrop. Ichthyol. 19 (2021).
Sato, L. Estudo das relações filogenéticas de Trichomycteridae (Teleostei, Siluriformes) com base em evidências cromossômicas e moleculares. (Universidade Estadual Paulista Júlio de Mesquita Filho - UNESP, 2007).
Ward, R. D., Zemlak, T. S., Innes, B. H., Last, P. R. & Hebert, P. D. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. B Biol. Sci. 360, 1847–1857 (2005).
Palumbi, S. R. Nucleic Acids II: The polymerase chain reaction. in Molecular Systematics, 2nd edition (eds. Hillis, D. M., Moritz, C. & Mable, B. K.) 205–247 (Sinauer, 1996).
Acknowledgements
The authors would like to express their gratitude to the members of the Ichthyology Research Group of the University of Antioquia for their invaluable support during the sampling campaigns. We are grateful to the specialists who reviewed the CIUA specimens: Henry. D. Agudelo-Zamora, Juan G. Albornoz-Garzón, Tiago P. Carvalho, Carlos A. García-Alzate, Miguel A. Hernández-Cortés, Flávio C. T. Lima, Nathan K. Lujan, Alejandro Méndez-López, and Andrea T. Thomaz. Additionally, we extend our sincere appreciation to Jorge E. García-Melo, Luz E. Ochoa, Gian C. Sánchez, Karen Lineke, Natalí Hernandez-Ciro, and Juliana López-Jiménez for their technical support. The study was funded through research agreements between the University of Antioquia and Empresas Públicas de Medellín (CT-2017-001714, CT-2021-00023-A3 and CW140036).
Author information
Authors and Affiliations
Contributions
L.F.J.S.; conceptualization, data curation, formal analysis, funding acquisition, methodology, organization of collecting trips, original investigation design, project administration, resources supervision, visualization, writing original draft, writing review, and editing. D.R.S.; conceptualization, data curation, formal analysis, methodology, organization of collecting trips, participation in most fish collecting trips, project administration, validation, visualization, writing original draft, writing review, and editing. J.G.O.P.; data curation, formal analysis, methodology, organization of collecting trips, participation in most fish collecting trips, validation, visualization, writing review, and editing. M.C.C.M.; data curation, formal analysis, methodology, participation in most fish collecting trips, validation, visualization, writing original draft, writing review, and editing. D.V.R.; data curation, organization of collecting trips, participation in most fish collecting trips, validation, writing original draft, writing review, and editing. A.F.G.; funding acquisition, organisation of collecting trips, project administration, resources supervision, writing review, and editing. J.L.L.L.; data curation, methodology, participation in most fish collecting trips, visualization, writing review, and editing. J.H.P.; data curation, organization of collecting trips, participation in most fish collecting trips, writing review and editing. V.M.M.R.; data curation, methodology, participation in most fish collecting trips. J.A.B.; data curation, methodology, participation in most fish collecting trips. M.M.E.; data curation, participation in most fish collecting trips. M.H.Z.; organization of collecting trips, project administration. L.J.G.M.; administration, resources supervision. O.C.N.; methodology. I.D.S.C.; data curation, methodology, writing review, and editing. C.D.; conceptualization, data curation, methodology, organization of collecting trips, project administration, validation, writing of the original draft, writing review, and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jiménez-Segura, L.F., Restrepo-Santamaria, D., Ospina-Pabón, J.G. et al. Fish databases for improving their conservation in Colombia. Sci Data 12, 262 (2025). https://doi.org/10.1038/s41597-024-04352-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-024-04352-3