Abstract
The understanding of glioma disease has significantly advanced through the application of genetic and molecular profiling techniques on brain tumour tissue. Molecular biomarkers have gained a crucial role in glioma diagnosis, driving groundbreaking changes in the disease classification as standardised by the 2016 and 2021 World Health Organisation (WHO) Classification of Tumours of the Central Nervous System. Recent insights from large-scale multi-omics databases, such as The Cancer Genome Atlas (TCGA), have enriched our comprehension of this cancer type. However, given the evolution of glioma classification, retrospective databases may contain outdated annotations, suboptimal for research. To address this issue, we propose two methods for updating the tumor classification of TCGA glioma samples according to the 2016 and 2021 WHO guidelines, through the integration of open-access curated molecular profiling data. Respectively, our Method-2016 and Method-2021 allowed for the diagnostic update of 98% and 87% of cases. The proposed reclassification pipelines, provided in R scripts, enable straightforward reproduction or customisation upon new WHO guideline releases.
Similar content being viewed by others
Introduction
Glioma is one of the most common form of primary central nervous system (CNS) malignant tumour in adults. These tumours typically exhibit diffuse and infiltrative behaviour, affecting the surrounding tissues and frequently causing disabling or fatal effects1.
Diffuse glioma pathologic diagnosis has traditionally been based on morphologic (histologic) features. Tumour population cell types were identified (astrocytes, oligodendrocytes or oligoastrocytic/mixed) and assigned a malignancy grade (glioma grade: II to IV), based on the degree of cell proliferation, florid microvascular proliferation and the presence or absence of necrosis2. These criteria lead to the recognition of different glioma types, namely: (i) astrocytic tumours — such as diffuse astrocytoma, anaplastic astrocytoma, and glioblastoma (GBM) —, (ii) oligodendroglial tumours — oligodendroglioma or anaplastic oligodendroglioma —, and (iii) oligoastrocytic (or “mixed”) tumours — oligoastrocytoma or anaplastic astrocytoma. Tumours were also described as Lower Grade Gliomas (LGG) when exhibiting lower proliferative activity (and, thus, grades II and III). This subcategory of glial tumours excluded the particular case of a diffuse astrocytoma, grade IV, denominated as GBM, which typically was associated with rapid pre- and postoperative disease evolution and fatal outcome3.
The accelerated knowledge gained on the biology of glioma has led to substantial changes in this disease’s classification over the years, breaking the century-old histogenetic classification of glial tumours4,5 as a result. Particularly, studies on molecular profiling of adult-type diffuse gliomas have established new insights for the identification of three molecular glioma groups6,7,8, first coined by a study by the Cancer Genome Atlas (TCGA)9: (i) “IDH-mutant, 1p/19q codeleted”, (ii) “IDH-mutant, 1p/19q non-codeleted”, and (iii) “IDH-wildtype”. IDH stands for isocitrate dehydrogenase genes IDH1 and IDH2, and 1p/19q codeletion (1p/19q codel) refers to the concomitant deletion of the 1p and the 19q chromosome arms (and 1p/19q non-codeletion (1p/19q non-codel) otherwise). The terms “IDH-mutant” and “IDH-wildtype” describe, respectively, the presence and absence of mutation in the IDH1 and/or IDH2 genes.
Given that the three above-mentioned tumour groups — defined by well-established, simple, and widely available markers — exhibited diverse clinical presentations and different survival patterns6, a revision of the gold-standard World Health Organization (WHO) classification of gliomas was pushed forward. Thus, with the release of the 2016 WHO Classification of Tumours of the CNS (WHO-2016)10, glioma classification was refined. For the first time, genetic and molecular profile information became integrated with histologic evaluation, therefore defining or “fine-tuning” the diagnosis of the disease.
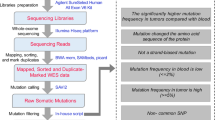
The tendency for reliance on molecular alterations continued to increase in the diagnosis and classification of gliomas. In 2021, the WHO Classification of Tumours of the CNS (WHO-2021) guidelines11 were published, greatly emphasising the role of genetic markers and molecular profiles of CNS tumours to determine the final diagnosis and to convey prognosis12. Fig. 1 summarises the highlights of the evolution of glioma classification as standardised by the WHO CNS guidelines, showcasing differences and novelties across the most recent versions.
Evolution of WHO CNS diagnostic criteria for glioma before and after WHO-2016 and WHO-2021 guidelines’ releases10,11. The scheme highlights the most prominent differences and similarities between the most recent versions of WHO CNS glioma classification10,11. Abbreviations: Astro, astrocytoma; codel, codeletion; IDH, isocitrate dehydrogenase; GBM, glioblastoma; mut, mutant/mutation; wt, wildtype; WHO, World Health Organization.
The insights gained from the molecular characterisation of gliomas have greatly benefited from the accessibility of publicly available datasets, such as those generated by the Cancer Genome Atlas (TCGA) program13. TCGA focuses on various cancer types, including gliomas, devoting specific projects to study LGG: the TCGA-LGG project, dedicated to glioma cases with WHO grades II and III14, and to study GBM: the TCGA-GBM project, aimed at GBM (WHO grade IV)15. However, despite TCGA data being continuously revised and managed by specialised entities16, its sample data collection ceased in 201317, and some glioma cases were assigned with histological subtypes that are currently known to be strongly discouraged in clinical practice — such as the “mixed glioma” (or oligoastrocytoma) subtype18. Hence, the diagnostic categories displayed in the datasets of TCGA-LGG and/or of TCGA-GBM projects may not be entirely consistent with the taxonomy suggested by WHO-2016 nor WHO-2021. Zakharova et al.19 recently proposed a procedure for the reclassification of TCGA glioma samples based on WHO-2021 guidelines, yet including evaluation of molecular features partly differing from the one brought forward with our study. A detailed comparison between our Method-2021 and the referred study is provided in the Supplementary Material A.
In this work, we propose two methodologies: Method-2016 and Method-2021, aiming at reevaluating diagnostic annotations of adult glioma samples and, through the integration of curated molecular profiling data and following the WHO-2016 and the WHO-2021 guidelines, assign them with updated tumour classes. Both methods were applied to the annotated samples belonging to the TCGA-LGG and TCGA-GBM datasets (hereafter referred to as the TCGA-PanGlioma dataset), by integrating correspondent molecular profiling data for each sample. To allow for a comparison between updated tumour classes resulting from the application of Method-2016 and of Method-2021, the end goal of our proposed methodologies was to categorise samples into updated and simplified tumour classes, i.e. “Astrocytoma”, “Oligodendroglioma” or “GBM”. This study focuses on the glioma types which in the WHO 2021 classification are members of the family “adult-type diffuse gliomas”; therefore, other types of glioma, such as, for instance, pediatric tumours, are out of the scope of this study.
Glioma classification: 2016 and 2021 WHO CNS guidelines
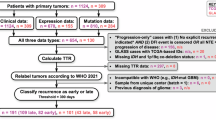
Here, we summarise the clinical diagnosis criteria for glioma disease as defined by the WHO-2016 and WHO-2021 guidelines, that served as the baseline for Method-2016 and Method-2021, respectively. Figure 2 simplifies both classification procedures for updating TCGA-PanGlioma tumour samples.
Simplified diagram illustrating the adult glioma classification procedures based on the (a) WHO-2016 and (b) WHO-2021 guidelines, the fundamental theory of the proposed Method-2016 and Method-2021, respectively. White rounded boxes represent the simplified glioma types: Astro (astrocytoma), Oligo (oligodendroglioma) and GBM (glioblastoma). In blue, the IDH mutation status can be mutant (IDHmut) or wildtype (IDHwt). In the scheme, “Others” refer to IDHwt patients who would require further evaluation to be completely classified. (a) TCGA glioma samples came histologically classified as Astro, Oligo, Mixed glioma (“Mix”) or GBM. Following the WHO-2016 guidelines, Method-2016 proposes samples to be further categorised according to molecular features provided by Ceccarelli et al.6. LGG samples with IDH mutation are classified as Astro in the absence of 1p/19q codeletion, or classified as Oligo in the presence of 1p/19q codeletion, while LGG samples without IDH mutation (wildtype: WT) are classified as other glioma types (“Others” in the scheme). GBM samples can be further categorised, according to their IDH mutation status, as either GBM IDHwt or GBM IDHmut. (b) Per WHO-2021 guidelines, Method-2021 proposes initial sample classification based on molecular profiles. IDH-mutant samples can be labelled as Astro or Oligo based on the absence or presence of 1p/19q codeletion, respectively. GBM IDHwt samples that exhibit particular histologic/genetic features are classified as GBM, otherwise considered as other glioma types (“Others” in the scheme). Abbreviations: Astro, Astrocytoma; EGFR, epidermal growth factor receptor; GBM, glioblastoma; IDH, isocitrate dehydrogenase; Mix, Mixed-glioma (i.e. oligoastrocytoma); mut, mutant; Oligo, oligodendroglioma; TERT, Telomerase reverse transcriptase; WHO, World Health Organization; wt/WT, wildtype; +7/10-, combined gain of entire chromosome 7 and loss of entire chromosome 10.
2016 WHO CNS guidelines
The WHO-2016 guidelines brought in a major restructuring of the diffuse glioma classification, officially introducing, for the first time, the concept of an “integrated” diagnosis, i.e. combined tissue-based histological and molecular diagnosis10. This newly introduced diagnostic objectivity aimed at establishing more biologically homogeneous and narrowly defined diagnostic entities than in prior classifications, to improve diagnostic accuracy and patient management, and convey more accurate determinations of prognosis and treatment response18.
Another clear example of the improvements deriving from the changes proposed in WHO-2016 relates to the diagnosis of oligoastrocytoma (or mixed-glioma) — a diagnostic category challenging to define and suffering from high interobserver discordance and inter-centre variability20. According to WHO-2016, this category became strongly discouraged in clinical practice, given that nearly all tumours with histological features suggesting both an astrocytic and an oligodendroglial components (i.e. “mixed” type) can be classified as either astrocytoma or oligodendroglioma using genetic testing (IDH mutation and 1p/19q codeletion statuses)18. As a result, both astrocytoma and oligodendroglioma cases became more homogeneously defined18. Rarely, IDH-wildtype mixed-gliomas can fall into different unconventional categories (represented in Fig. 2a as “Others”).
Thus, regarding glioma classification, WHO grade II diffuse astrocytomas, WHO grade III anaplastic astrocytomas and GBM became divided and further categorised through evaluation of IDH mutation status and/or 1p/19q codeletion. Thus, a tumour would be categorised as IDH-mutant if exhibiting an IDH mutation or, on the contrary, as IDH-wildtype. The designation of NOS (Not Otherwise Specified) became reserved for tumours for which full IDH evaluation could not be performed. The final diagnostic nomenclature included both histologic and molecular profiling evidence (example of a tumour entity: “Diffuse astrocytoma, IDH-mutant”), and the tumour grading is predetermined by the diagnosed tumour entity18.
However, the simultaneous consideration of histological and molecular features has led to the emergence of tumour groups that challenge established categorisation into such distinct entities. Thus, WHO-2016 criteria may potentially result in uncommon diagnoses, in which case it would be needed careful further evaluation to prevent misdiagnosis (e.g. “diffuse astrocytoma, IDH-wildtype”; or “anaplastic astrocytoma, IDH-wildtype”)18. Also, the new criteria have the potential to raise discordant tumour-type results. In such cases, a “two-layered” diagnosis would be needed, reporting findings on genotype and histological phenotypes18. As an illustrative example, let us consider a case of a diffuse glioma with astrocytic phenotype while exhibiting IDH mutation and 1p/19q codeletion. This case would require the assignment of two diagnostic annotations: “diffuse astrocytoma, IDH-mutant” (based on histological phenotype) and “oligodendroglioma, IDH-mutant and 1p/19q codeleted” (based on genotype).
2021 WHO CNS guidelines
The 2021 release of the WHO Classification of Tumours of the CNS11 extends and enhances the groundwork established by the WHO-2016. It introduces novel approaches to CNS tumour nomenclature and grading, highlighting the significance of molecular biomarkers for an “integrated diagnosis”, while notably greatly simplifying the classification of adult-type diffuse gliomas. The prominent updates in this edition, regarding glioma, include the formal differentiation between paediatric and adult-type tumours, as well as a novel approach to grading. Regarding the latter, tumours became graded within their specific type, rather than being graded across different tumour types, and the new WHO CNS grade scale uses Arabic numerals from 1 to 4, replacing the previous use of Roman numerals.
While in WHO-2016 diffuse gliomas were highly divided (a consequence of different tumour grades leading to different tumour names/entities), in WHO-2021, adult-type diffuse gliomas are composed of only three types. Namely, (i) “Oligodendroglioma, IDH-mutant and 1p/19q-codeleted” (grades 2 or 3) in the case of a diffuse glioma exhibiting combined IDH mutation and 1p/19q codeletion; (ii) “Astrocytoma, IDH-mutant” (grades 2, 3 and 4) for an IDH-mutant diffuse astrocytic tumour without 1p/19q codeletion; and (iii) “Glioblastoma, IDH-wildtype” (grade 4). The latter diagnosis is assigned in the setting of an IDH-wildtype diffuse and astrocytic glioma in the presence of certain genetic parameters (TERT — Telomerase reverse transcriptase — promoter mutation, or EGFR (epidermal growth factor receptor) gene amplification, or combined gain of entire chromosome 7 and loss of entire chromosome 10 (+7/−10)), or if exhibiting histologic hallmarks such as microvascular proliferation or necrosis11. In the rare occasion that none of the referred genetic or histologic features is present in an IDH-wildtype diffuse and astrocytic glioma, such results do not allow for a WHO diagnosis, and other types of tumour families are considered (e.g. paediatric tumour-type) (represented as “Others” in Fig. 2b)11.
Results
The implementation of Method-2016 and of Method-2021 to the TCGA-PanGlioma dataset (composed of the sum of TCGA-GBM and TCGA-LGG projects datasets) has allowed for the assignment of updated classes for the majority of evaluated adult glioma cases (Table 1).
Method-2016 was employed to the TCGA-LGG dataset, comprised of 130 mixed glioma samples, allowing for the reclassification of 114 cases: 78 astrocytomas and 36 oligodendrogliomas — the remaining samples (n = 16) were unable to be classified (lack of information to assign a WHO-diagnosis). This method further characterised 458 out of 595 GBM samples from TCGA-GBM according to their IDH gene mutation status (“Glioblastoma, IDH-mutant” (n = 35), “Glioblastoma, IDH-wildtype” (n = 423) and “Glioblastoma, NOS (n = 137)). Method-2016 has produced an updated version of 1094 out of 1110 samples from the TCGA-PanGlioma dataset, specifically, 272 astrocytomas, 227 oligodendrogliomas and 595 GBM (Table 1, last column).
Method-2021 has evaluated a total of 1110 glioma samples from the TCGA-PanGlioma dataset, generating the final dataset composed of 959 cases: 282 astrocytomas, 171 oligodendrogliomas and 506 GBM. A total of 151 samples lacked information to be reclassified with this methodology (Table 1, last row).
In Table 1, a comparative analysis is presented, outlining the diagnostic classifications resulting from Method-2016 and Method-2021. In the initial row of Table 1, we can assess that among the 285 samples classified as astrocytoma using Method-2016, 208 maintained this diagnosis with Method-2021, five cases transitioned to oligodendroglioma, 41 samples shifted from astrocytoma to GBM, and 18 could not be classified with Method-2021 due to a lack of molecular information. The cross-table emphasises the presence of 41 samples changing from oligodendroglioma to astrocytoma, along with five cases converting from astrocytoma to oligodendroglioma.
It is noteworthy that, in the transition from 2016 to 2021 guidelines’ criteria, samples can exhibit shifts from LGG to GBM group, and vice versa. Specifically, 41 samples classified as 2016-astrocytoma (i.e. classified as astrocytoma by Method-2016), and 12 samples classified as 2016-oligodendroglioma are now respectively classified as 2021-GBM (i.e. classified as GBM by Method-2021), while 2 cases classified as 2016 GBM would now belong to oligodendroglioma. In addition, we identified 33 cases classified as GBM in 2016 that are now diagnosed as astrocytoma. These represent high-grade astrocytomas, newly defined in the 2021 WHO classification as “Astrocytoma, IDH-mutant, grade 4”.
Given the retrospective nature of the data, an analysis of the omics data from TCGA samples was conducted to validate the updated labels. Our findings based on several methods21,22,23 indicate improved performance in omics studies when using labels from Method-2021, suggesting a better alignment between diagnostic labels and omics profiles. Details and results of this validation procedure are provided in the Supplementary Material B.
Discussion
CNS tumour classification has been evolving drastically over the years. The stage of understanding of the neuropathology field at a particular time is reflected in every new release of WHO CNS guidelines, providing worldwide practical guidance to pathologists and neuro-oncology specialists and new curated knowledge for the classification of tumour entities.
We have integrated tumour-type annotations of LGG and GBM adult cases from TCGA with curated molecular profile data, to reevaluate them and reclassify the tumoural samples according to the WHO guidelines of 2016 and 2021 with Method-2016 and Method-2021, respectively. Our reclassification pipelines are easily reproducible with the R scripts available at https://github.com/sysbiomed/MONET GitHub repository (Code Availability Section).
Overall, the outcome of the implementation of Method-2016 on TCGA-PanGlioma samples allowed for the reclassification of 114 out of 130 mixed glioma samples (Figure SB1), a diagnostic category that became strongly discouraged in clinical practice. Method-2016 was designed to adopt a conservative approach in reevaluating the TCGA diagnosis of astrocytoma, oligodendroglioma and GBM samples, solely relying on prior TCGA histological classes. The Method exclusively assigned a univocal, simplified glioma type label correspondent to the retrospective TCGA diagnosis, ensuring the preservation of the histologic information. This choice prevents assigning new labels for samples eventually presenting contradictory molecular and histological features. According to WHO-2016, these particular cases would require careful examination, resulting in a) two-layered diagnosis or b) tumour entities that were not yet distinguished as solid disease entities at the time (i.e. provisional)18, or c) uncommon diagnosis (e.g. “diffuse astrocytoma, IDH-wildtype” or “anaplastic astrocytoma, IDH-wildtype”)10. The patients that would be assigned with the two-layered diagnosis according to WHO-2016 can be identified in Table 1, represented by the cases which, according to WHO-2021, changed their WHO-2016 glioma type from astrocytoma to oligodendroglioma (n = 5), and from oligodendroglioma to astrocytoma (n = 41). This transition highlights the discordance between histological and molecular features that WHO-2016 classification could give rise to, e.g. samples that would be classified as astrocytoma through histological evaluation, while exhibiting molecular features of oligodendroglioma (IDH-mutant and 1p/19q codeleted). Even though the classification provided by Method-2016 does not take into account the complete diagnosis, our procedure avoids introducing uncertainty into the classification for those cases that would need further evaluation. Moreover, Method-2016 allows for potential retrospective analyses regarding the original histological information provided by TCGA, for direct comparison between the two most recent guidelines, as well as the opportunity to look into cases that would have different classifications according to WHO-2016 and WHO-2021. The complexity of this diagnosis has been reduced with the new WHO-2021 guidelines, which assign a unique label for such cases, mainly based on genetic markers and molecular profiles. Specifically, with WHO-2021, the new concept of “integrated diagnosis” has been emphasised, combining histological and molecular features to define a univocal final diagnosis, including the tumour grade.
In this work’s Method-2016 and Method-2021, tumour grades were not considered when reevaluating TCGA-PanGlioma cases due to several considerations. Over the latest guideline revisions, the grading process had undergone significant changes, passing from being tumour entity-specific to within-tumour-type, and being influenced by histological features, molecular biomarkers, and clinical context11,24. Also, the data constraints of this retrospective study impair the tumour grade re-evaluation for the complete TCGA-PanGlioma dataset, as many samples would require specific revision of the histopathological features, which are not available. Furthermore, nowadays, the necessity of assigning a grade to every case is still under debate as it may be a source of confusion in clinical care12. In fact, given the great impact that modern molecular-based treatments could have on patients’ prognosis, currently, CNS tumour grades may no longer reflect the expected clinical-biological behaviour25.
Finally, although the integration of molecular profile data with TCGA diagnostic annotation allowed us to establish WHO CNS diagnostic classes for the great majority of glioma cases, there were samples unable to be classified. While with Method-2016 only 1% of all the cases could not be updated, with Method-2021 the number of unclassified samples substantially increased to 13% (n = 151). This has occurred due to: (i) overall unavailable IDH status information (n = 120), (ii) lack of 1p/19q codeletion status data for IDH-mutant cases manually searched on GDC Data Portal16 (n = 2), and (iii) IDH-wildtype cases requiring additional information to acquire the diagnosis of “Glioblastoma, IDH-wildtype” (n = 29) — the latter happens since, for those specific cases, we had no access to one of the three molecular markers (EGFR amplification) that can define this diagnosis.
Despite the retrospective nature of the data from TCGA, the integration of molecular profiling data enabled the update of 1094 (98%) and 959 (87%) glioma types following the latest WHO guidelines of 2016 and 2021, respectively. Moreover, the analysis carried with TCGA-PanGlioma omics data and the newly assigned diagnostic labels allowed for the proposed methods’ validation, supporting the improvement of glioma diagnosis through their genetic and molecular characterisation as indicated by the latest WHO CNS classification guidelines.
In identifying cases with outdated diagnostic labels, such as “mixed glioma”, and emphasising the potential for substantial changes in their glioma type and/or their broader categories (LGG or GBM) based on curated genetic and molecular data by Ceccarelli et al.6, our study prompts further exploration of TCGA-PanGlioma cases. These deviations can have significant implications for future bioinformatic and statistical analyses dedicated to studying and characterising different glioma cases. We anticipate that this reclassification pipeline for glioma types, according to the latest WHO CNS guidelines, will provide new pathways for the scientific community, facilitating analyses of glioma disease within genomic databases, and ultimately contributing to the well-being of patients affected by this condition.
Methods
For reclassification of TCGA-PanGlioma cases according to the WHO-2016 and -2021 guidelines, we have developed two methods: Method-2016 and Method-2021, respectively. Both methods require, to a greater or lesser extent, processing the diagnostic information specified on TCGA-PanGlioma clinical data (class and/or histological type) and the integrated molecular information (genetic and/or molecular profiling data) provided in the study of Ceccarelli et al.6. It is noteworthy that the considered cases in this study are those that in the WHO-2021 would belong to the category of “adult-type diffuse gliomas”.
TCGA pan-glioma patient cohort characteristics
The latest versions of TCGA-LGG and TCGA-GBM cohorts are composed of 515 and 595 patients, respectively, together composing the TCGA-PanGlioma dataset with a total of 1110 glioma cases. The TCGA-LGG project targeted the study of untreated lower grade gliomas, consisting of grades II and III, including 194 astrocytomas, 191 oligodendrogliomas, and 130 oligoastrocytomas/mixed-gliomas9,14. TCGA-GBM project is composed of samples selected based on an anatomic pathology diagnosis of GBM (equivalent to an astrocytoma WHO grade IV)8. This project started in 200815 and it was expanded in 20137, characterising 595 GBM tumours.
Molecular profiling data of tcga-panglioma samples
The molecular profiling data is provided by the study of Ceccarelli et al.6 (n = 960 samples). The study performed a comprehensive multi-platform genomic analysis of adult diffuse II, III and IV glioma cases from TCGA, including DNA methylation profile and whole-genome sequencing data analysis. Among other molecular features, it includes information on IDH mutation and 1p/19q co-deletion statuses, as well as TERT promoter mutations and the presence of +7/-10. Samples with missing information about IDH1 and IDH2 genes statuses (n = 140) have been manually searched on Genomic Data Commons (GDC) Data Portal16, the platform that currently houses and manages TCGA data.
Data extraction and integration
Clinical data from TCGA-LGG and TCGA-GBM projects, including the assigned diagnosis of each tumoural sample (one per patient), was acquired through the getFirehoseData function from RTCGAToolbox package26 from R software (version 3.5.1, https://www.r-project.org). We have downloaded the latest Firehose data release available (2016-01-28), as it coincides with the latest clinical data available at the GDC Data Portal16.
The molecular profiling data, curated by Ceccarelli et al.6, was downloaded through the TCGABiolinks R package27,28,29, using the “PanCancerAtlas_subtypes” function and the keywords “lgg” and “gbm”. This data can also be accessed through the supplementary materials provided in the mentioned article. Among the samples included in this study, 140 samples had no information about IDH mutation status. For such cases, we have manually searched for IDH mutation status on GDC Data Portal16, finding information for 20 samples.
We have merged the two datasets (diagnostic annotation from TCGA and molecular profiling) by the sample’s TCGA-specific barcode, which is composed of a collection of identifiers. We first selected TCGA barcodes referring to primary tumours (sample code “01”), and then reduced the initial TCGA barcode in order to keep only the first two fields (tissue source site and participant, e.g. TCGA-02-0001). This data integration procedure led to a final TCGA-PanGlioma dataset comprising 1110 samples (515 TCGA-LGG and 595 TCGA-GBM). Unmatched samples (n = 12) were excluded, as they were only present in the molecular profiling data of Ceccarelli et al.6, and not in the TCGA-LGG and/or on the TCGA-GBM latest data release.
Method-2016
Method-2016 aims to update the diagnostic annotations of TCGA-PanGlioma cases in accordance with the WHO-2016 (pipeline in Fig. 3a). It focuses on the reclassification of mixed glioma (or oligoastrocytoma) samples using IDH-mutation and 1p/19q codeletion statuses. GBM samples were further categorised according to IDH-mutation status.
Scheme summarising the classification pipeline of (a) Method-2016 and (b) Method-2021, with the characteristics of their intermediate datasets, and (c) reporting the final dimensions of the updated elements of the simplified adult glioma classes. Each step indicates the number of samples involved in the classification procedure. Abbreviations: Astro, Astrocytoma; 7 + /10−, concomitant chromosome 7 gain/10 loss; GBM, glioblastoma; Histo, Histologic features; IDH, isocitrate dehydrogenase; LGG, Low Grade Glioma; Mix, Mixed-glioma (i.e. oligoastrocytoma); mut, mutant; NA, not available; Oligo, oligodendroglioma; TERT, Telomerase reverse transcriptase; WHO, World Health Organization; wt, wildtype.
Samples with TCGA diagnostic annotation of “Anaplastic Astrocytoma”, “Astrocytoma, NOS”, “Anaplastic Oligodendroglioma” or “Oligodendroglioma, NOS” were assigned with a corresponding simplified tumour type label, i.e. either “Astrocytoma” or “Oligodendroglioma”, maintaining the retrospective TCGA diagnosis, and securing the histologic information (see Discussion for more details).
-
i).
Reclassification of oligoastrocytoma (mixed glioma) samples
Step 1: Select patients with a diagnosis of “mixed glioma” (or, equivalently, “oligoastrocytoma”).
Step 2: Consult the IDH status and 1p/19q codeletion status of each patient sample.
Case 1) If the patient is “IDH mutant-non-codel”, it is reclassified as “Astrocytoma” (Diffuse astrocytoma, IDH-mutant).
Case 2) If the patient is “IDH mutant-codel”, it is assigned as “Oligodendroglioma” (Oligodendroglioma, IDH-mutant and 1p/19q codeleted).
Case 3) If the patient is “IDH-wildtype”, it is identified as “Unclassified”.
-
ii).
Further categorisation of GBM samples
Similarly, we have selected patients with a diagnostic category of GBM. We have evaluated these cases with the integrated information on IDH status (wildtype or mutant) from the molecular profiling dataset and assigned them new diagnostic types of either “Glioblastoma, IDH-wildtype” or “Glioblastoma, IDH-mutant”. If the sample lacks information on the IDH status, it is given the diagnosis of “Glioblastoma, NOS”.
Method-2021
With Method-2021 we reclassified each TCGA-PanGlioma case following the WHO-2021 guidelines (pipeline schematised in Fig. 3b). We mainly used these patients’ molecular information on “IDH-mutation” and “1p/19q co-deletion” statuses, disregarding the previous clinical annotations from TCGA (derived from histological features). The unique exception regards IDH-wildtype samples, which need additional information to be classified as “Glioblastoma”. In these cases, we considered the status of TERT mutation and the presence of +7/-10, as well as the previous TCGA diagnostic annotation. Indeed, the histologic hallmarks used to identify GBM did not change over the years, therefore being classified as “glioblastoma” on TCGA indicates the presence of necrosis or microvascular proliferation. This information, coupled with IDH-wildtype, is sufficient to classify these samples as GBM.
We have considered all samples from the TCGA-PanGlioma dataset. Adopting the WHO-2021 guidelines, we have integrated the clinical and molecular data of 1100 TCGA glioma cases, and reassigned them to the corresponding glioma type according to the following criteria:
Case 1) If the sample has IDH mutation:
Case 1.1) If it has 1p/19q codeletion, it is classified as “Oligodendroglioma” (“Oligodendroglioma, IDH-mutant and 1p/19q codeleted”).
Case 1.2) If the sample does not exhibit 1p/19q codeletion, it is classified as “Astrocytoma” (“Astrocytoma, IDH-mutant”).
Case 2) If the sample is IDH-wildtype:
Case 2.1) If the TCGA diagnostic annotation was of “glioblastoma” or if a TERT promoter mutation is present or exhibits +7/−10, the sample is classified as “GBM” (“Glioblastoma, IDH-wildtype”).
Case 2.2) Otherwise, we label the sample as “Unclassified”.
Regarding the samples we integrated manually from TCGA (n = 20), 18 belonged to the TCGA-GBM project and were IDH-wildtype, thus diagnosed as “Glioblastoma, IDH-wildtype” according to Case 2. The remaining 2 samples were IDH-mutant from the TCGA-GBM project but lacked information on 1p/19q codeletion status to be reclassified according to Case 1. These patients were not updated (we could only assess that these 2 samples would transition from GBM to LGG groups).
Given the high complexity of the nomenclature of glioma entities in WHO-2016 and WHO-2021 guidelines, our main goal is to provide a simplified label in order to divide samples into the three main tumour classes, namely “Astrocytoma”, “Oligodendroglioma” and “GBM”. Samples that cannot be classified due to lack of information have been labelled as “Unclassified”. More detailed annotations (in accordance with WHO CNS taxonomy) are available in the files “Output1”, “Output2” and “Output3” at https://github.com/sysbiomed/MONET and figshare30,31,32 .
Data availability
The updated diagnostic labels provided by the presented study are publicly available at the GitHub repository https://github.com/sysbiomed/MONET and figshare30,31,32. These final outputs are organised in three files described in the following item.
• “Matrix_WHO2016.csv” (Output 1) resumes the sample classification provided by Method-2016. This 1110 × 4 matrix reports, for each sample, the corresponding histological type currently available on TCGA (“TCGA-histological.type” column), the exact type nomenclature assigned by our method, in accordance with the 2016 WHO CNS taxonomy (“classification.2016_complete.labels” column), as well as the simplified name we attributed (“classification.2016_simplified.labels” column).
The WHO-2016 nomenclature terms considered were:
- diffuse _astrocytoma_IDHmut (if astrocytoma or mixed glioma with IDH mutation)
- oligodendroglioma_IDHmut_1p19qcodel (if oligodendroglioma or mixed-glioma with IDH mutation and 1p/19q codeletion)
- diffuse_astrocytoma_IDHwt (if astrocytoma with IDHwt)
- oligodendroglioma_NOS (if oligodendroglioma with either IDHwt or IDHmut but 1p/19q non-codel, or missing information on IDH mutation and 1p/19q codeletion status)
- diffuse_astrocytoma_NOS (if astrocytoma with missing information on IDH status)
- glioblastoma_IDHwt (if GBM with IDHwt)
- glioblastoma_IDHmut (if GBM with IDH mutation)
- glioblastoma_NOS (if GBM with missing information on IDH status)
- unclassified (if mixed glioma with IDHwt)
• “Matrix_WHO2021.csv” (Output 2) resumes the sample classification provided by Method-2021. This 1110 × 5 matrix reports, for each sample, the corresponding histological type currently available on TCGA (“TCGA-histological.type” column), the exact type nomenclature assigned by our method, in accordance with the 2021 WHO CNS taxonomy (“classification.2021_complete.labels” column), as well as the simplified name we attributed (“classification.2021_simplified.labels” column). The WHO-2021 nomenclature terms considered were: “astrocytoma_IDHmut” (if IDHmut without 1p/19q codel), “olgodendroglioma_IDHmut_1p19qcodel” (if IDHmut with 1p/19q codel), “glioblastoma_IDHwt” (if IDHwt + histological or genetic hallmarks) and “unclassified” (in case of missing information).
• “SIMPLIFIED_CLASSIFICATION_TCGA_2016_2021.csv” (Output 3), which compares, for each sample, the current TCGA histological type classes (“TCGA.histological.type” column), with the simplified label we provide by applying Method-2016 (“classification.2016” column) and Method-2021 (“classification.2021” column).
Code availability
The proposed Method-2016 and Method-2021 are publicly available at the GitHub repository https://github.com/sysbiomed/MONET. Table 2 provides a list of scripts and files included in these folders.
References
American Cancer Society. Brain and Spinal Cord Tumors in Adults. http://www.cancer.org/Cancer/BrainCNSTumorsinAdults/DetailedGuide/brain-and-spinal-cord-tumors-in-adults-what-are-brain-spinal-tumors (Accessed: November 2, 2022).
Mesfin, F. B., & Al-Dhahir, M. A. Gliomas. In StatPearls. StatPearls Publishing (2022).
Louis, D. N. et al. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol 114, 97–109, https://doi.org/10.1007/s00401-007-0243-4 (2007).
Perez, A. & Huse, J. T. The Evolving Classification of Diffuse Gliomas: World Health Organization Updates for 2021. Curr Neurol Neurosci Rep 21, 67, https://doi.org/10.1007/s11910-021-01153-8 (2021).
MacKenzie, D. J. A Classification of the Tumours of the Glioma Group on a Histogenetic Basis With a Correlated Study of Prognosis. Canadian Medical Association Journal 16(7), 872 (1926).
Ceccarelli, M. et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 164(3), 550–563, https://doi.org/10.1016/j.cell.2015.12.028 (2016).
Brennan, C. W. et al. The somatic genomic landscape of glioblastoma. Cell 155(2), 462–477, https://doi.org/10.1016/j.cell.2013.09.034 (2013).
Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455(7216), 1061–1068, https://doi.org/10.1038/nature07385 (2008).
Cancer Genome Atlas Research Network et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. The New England journal of medicine 372(26), 2481–2498, https://doi.org/10.1056/NEJMoa1402121 (2015).
Louis, D. N., Ohgaki, H., Wiestler, O. D., Cavenee, W. K. World Health Organization Classification of Tumours of the Central Nervous System. 4th ed., updated ed. Lyon: International Agency for Research on Cancer (2016).
WHO Classification of Tumours Editorial Board World Health Organization Classification of Tumours of the Central Nervous System. 5th ed. Lyon: International Agency for Research on Cancer (2021).
Louis, D. N. et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro-oncology 23(8), 1231–1251, https://doi.org/10.1093/neuonc/noab106 (2021).
Tomczak, K., Czerwińska, P. & Wiznerowicz, M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemporary oncology (Poznan, Poland) 19(1A), A68–A77, https://doi.org/10.5114/wo.2014.47136 (2015).
The Cancer Genome Atlas - Lower Grade Glioma study. National Cancer Institute. Available at: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/studied-cancers/glioma (Accessed: October 3, 2022).
The Cancer Genome Atlas - Glioblastoma Multiforme Study. National Cancer Institute. Available at: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/studied-cancers/glioblastoma (Accessed: October 3, 2022).
Grossman, R. L. et al. Toward a Shared Vision for Cancer Genomic Data. The New England journal of medicine 375(12), 1109–1112, https://doi.org/10.1056/NEJMp1607591 (2016).
The Cancer Genome Atlas - Timeline and Milestones. National Cancer Institute. Available at: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/history/timeline (Accessed: October 4, 2022).
Louis, D. N. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta neuropathologica 131(6), 803–820, https://doi.org/10.1007/s00401-016-1545-1 (2016).
Zakharova, G. et al. Reclassification of TCGA Diffuse Glioma Profiles Linked to Transcriptomic, Epigenetic, Genomic and Clinical Data, According to the 2021 WHO CNS Tumor Classification. International journal of molecular sciences 24(1), 157, https://doi.org/10.3390/ijms24010157 (2023).
Van den Bent, M. J. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician’s perspective. Acta neuropathologica 120(3), 297–304, https://doi.org/10.1007/s00401-010-0725-7 (2010).
Coletti, R., Mendonça, M. L., Vinga, S., Lopes, M. B. Inferring diagnostic and prognostic gene expression signatures across who glioma classifications: A network-based approach. Bioinf Biol Insights, 18. https://doi.org/10.1177/11779322241271535 (2024).
Vieira, F.G., Bispo, R., Lopes, M. B. Integration of Multi-Omics Data for the Classification of Glioma Types and Identification of Novel Biomarkers. Bioinf Biol Insights, 18. https://doi.org/10.1177/11779322241249563 (2024).
McInnes, L., Healy, J. and Melville, J. UMAP: Uniform manifold approximation and projection for dimension reduction, arXiv.org. https://doi.org/10.48550/arXiv.1802.03426 (2020).
Louis, D. N. et al. cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol 30, 844–856, https://doi.org/10.1111/bpa.12832 (2020).
von Deimling, A., Ono, T., Shirahata, M. & Louis, D. N. Grading of Diffuse Astrocytic Gliomas: A Review of Studies Before and After the Advent of IDH Testing. Seminars in neurology 38(1), 19–23, https://doi.org/10.1055/s-0038-1636430 (2018).
Samur, M. K. RTCGAToolbox: a new tool for exporting TCGA Firehose data. PloS one 9(9), e106397, https://doi.org/10.1371/journal.pone.0106397 (2014).
Colaprico, A. et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic acids research 44(8), e71, https://doi.org/10.1093/nar/gkv1507 (2016).
Silva, T. C. et al. TCGA Workflow: Analyze cancer genomics and epigenomics data using Bioconductor packages. F1000Research 5, 1542, https://doi.org/10.12688/f1000research.8923 (2016).
Mounir, M. et al. New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS computational biology 15(3), e1006701, https://doi.org/10.1371/journal.pcbi.1006701 (2019).
Mendonça, M. L. et al. Updated TCGA glioma classification through integration of molecular data following the 2016 WHO guidelines. Figshare https://doi.org/10.6084/m9.figshare.28173470.v1 (2025).
Mendonça, M. L. et al. Updated TCGA glioma classification through integration of molecular data following the 2021 WHO guidelines. Figshare https://doi.org/10.6084/m9.figshare.28173476.v1 (2025).
Mendonça, M. L. et al. Comparison of the current TCGA histological type classes with the simplified label provided by Method-2016 and Method-2021. Figshare https://doi.org/10.6084/m9.figshare.28173473.v1 (2025).
Acknowledgements
This work was supported by national funds through Fundação para a Ciência e a Tecnologia (FCT) with references CEECINST/00102/2018, 2021.02600.CEECIND, PD/BDE/143154/2019, COVID/BDE/153298/2023, UIDB/00297/2020 and UIDP/00297/2020 (NOVA Math, Center for Mathematics and Applications), UIDB/04516/2020 (NOVA LINCS), UIDB/50026/2020 and UIDP/50026/2020 (ICVS), UIDB/50021/2020 (INESC-ID), UIDB/50022/2020 (IDMEC), and the research project “MONET – Multi-omic networks in gliomas” (PTDC/CCI-BIO/4180/2020). This work was also supported by the project NORTE-01-0145-FEDER-000055, supported by Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF). This project has received funding from the European Union’s Horizon 2020 research and innovation program under Grant Agreement No. 951970 (OLISSIPO project). Input Datasets: The results shown here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Author information
Authors and Affiliations
Contributions
M.L.M. and R.C. are co-first authors in this study. All authors conceptualised the study. M.L.M. designed the methodology and its computational application, conducted research, and collected open-source data. R.C. implemented the methodology, deployed computer codes, analysed the data, performed data curation and technical validation. M.L.M. and R.C. wrote the original draft, interpreted the results, reviewed and edited the visualisation content. C.S.G., E.P.M. and B.M.C. validated the theoretical background, methodology and outcomes from the biological point of view. B.M.C., S.V. and M.B.L. supervised the study, administered the project and acquired funding supporting this project. All authors reviewed and edited the published work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
de Mendonça, M.L., Coletti, R., Gonçalves, C.S. et al. Updating TCGA glioma classification through integration of molecular data following the latest WHO guidelines. Sci Data 12, 935 (2025). https://doi.org/10.1038/s41597-025-05117-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05117-2
This article is cited by
-
Prognostic value of LPAR1 expression and methylation in low-grade gliomas: a meta-analysis of TCGA and CGGA datasets and functional validation
BMC Cancer (2025)
-
Deep learning model outperforms traditional models in clinical data-based prognostic prediction for adult-type diffuse glioma
Discover Oncology (2025)