Abstract
Although multiple single-cell RNA sequencing (scRNA-seq) based studies have explored rheumatoid arthritis (RA), most have focused on synovial tissue, limiting the ability to perform paired pre- and post-treatment comparisons and to elucidate dynamic changes. Moreover, the single-cell impact of TNF-α and JAK inhibitors on RA remains poorly understood, hindering effects toward precision medicine for RA. Here, we present a comprehensive scRNA-seq analysis of synovial fluid samples of RA patients treated with tofacitinib or adalimumab, both pre- and post-treatment. Following stringent quality control, we analyzed 100,387 high-quality cells, providing an in-depth characterization of cell composition, molecular signaling pathways, and functional properties. This rich dataset offers a valuable resource to address diverse research questions, from uncovering the mechanisms of action of TNF-α and JAK inhibitors, to identifying novel therapeutic targets for RA, and advancing our understanding of the pathogenesis of inflammatory diseases.
Similar content being viewed by others
Background & Summary
Rheumatoid arthritis (RA) is a chronic inflammatory autoimmune disease that typically presents as symmetrical involvement of small joints, characterized by joint swelling, ongoing bone destruction, and permanent joint deformity1,2,3. Current clinical treatments of RA include glucocorticosteroids, non-steroidal anti-inflammatory drugs and disease-modifying antirheumatic drugs (DMARDs). DMARDs, which are further classified into conventional synthetic DMARDs (csDMARDs), biological DMARDs (bDMARDs), and targeted synthetic DMARDs (tsDMARDs), play a critical role in inhibiting bone erosion and joint destruction. Among these, b/tsDMARDs target specific inflammatory molecules, offering rapid and effective symptom relief. Although RA remains incurable, 20–40% of RA patients achieve ≥ 70% improvement criteria based on American College of Rheumatology (ACR70) criteria after six months of csDMARDs treatment, such as methotrexate (MTX). For patients who are insufficient responders to MTX, approximately 20% achieve similar improvements with bDMARDs (e.g., adalimumab, targeting TNF-α) or tsDMARDs (e.g., tofacitinib, targeting the downstream JAK-STAT pathway)4,5,6,7,8. Current clinical guidelines recommend combining b/tsDMARDs with csDMARDs for patients who show inadequate response to csDMARDs9. However, these guidelines lack detailed prioritization for the selection between bDMARDs and tsDMARDs, offering tailored recommendations only for specific patient groups. For instance, advises against the use of TNF inhibitors and instead recommends other bDMARDs or tsDMARDs for RA patients with heart failure10. Similarly, the European Medicines Agency (EMA) recommends tofacitinib only for patients aged ≥ 65 years, smokers, or those with cardiovascular or malignancy risk factors, and only when no suitable alternatives are available11. Despite these recommendations, a significant proportion of RA patients in clinical practice do not have contraindications such as cancer, heart failure, or cardiovascular risks. For this broader population, the selection of the most appropriate b/tsDMARD and the development of precise treatment strategies remain inadequately supported by reliable evidence. Understanding the changes in cellular immune profiles before and after treatment with b/tsDMARDs is thus critical for optimizing drug selection and advancing precision medicine in RA management.
Previous studies investigating the cellular immune response in RA have primarily focus on synovial tissue (ST). While ST is a key target organ in RA, its inaccessibility outside surgical settings limits its utility for routine diagnosis or treatment evaluation. Synovial fluid (SF), in contrast, is co-located with ST in the joint cavity’s inflammatory environment and is readily obtained through arthrocentesis. Previous studies have identified potential biomarkers in SF, such as C-reactive protein and calcium granule binding protein12,13, while flow cytometry has revealed imbalances in macrophage subpopulations. However, these methods lack the resolution to comprehensively characterize the transcriptional, functional diversity, and cell-cell communications of SF cells in RA. Single-cell RNA sequencing (scRNA-seq) addresses this gap by enabling high-throughput transcriptome profiling at single-cell resolution, offering a deeper understanding of cellular heterogeneity and intercellular interactions of immune microenvironment in RA.
In this study, we collected SF samples from RA patients pre- and post-treated with TNF-α/JAK inhibitors and constructed a single-cell atlas of RA SF. This dataset offers a valuable resource for identifying therapeutic targets, understanding the mechanisms of TNF-α/JAK inhibitors (i.e., adalimumab and tofacitinib), and exploring the pathogenesis of inflammatory diseases.
Methods
Ethical statement
This study was approved by the Ethics Committee of West China Hospital, Sichuan University (Approval Number: 2020-1151). All participants provided informed consent for the sharing of their transcriptomic data and were made aware of any associated risks. The open access use of these data was permitted by the Ministry of Science and Technology of China (Registration Number: 2022BAT2228).
Participant information
In our study, nine patients who met the 2010 revised criteria14 for RA established by the American College of Rheumatology were prospectively and randomly enrolled from department of rheumatology and immunology, West China Hospital, Sichuan University. Informed consent was obtained from all participants. At baseline, all patients were required to exhibited high or moderate disease activity and to have an inadequate response to methotrexate. In addition, these patients were required not received glucocorticoids or any bDMARDs or tsDMARDs within 3 months prior to enrollment. Clinical and demographic details are provided in Supplementary Table 1-2. To comprehensively define the transcriptional atlas of SF cells from RA patients, who are MTX insufficient responders, we performed scRNA-seq on nine paired samples from RA patients before and after one month of treatment with TNF-α or JAK inhibitors (referred to as RA-BT and RA-AT, respectively). Specifically, nine enrolled RA patients were treated with either adalimumab (n = 5, 40 mg every 2 weeks) or tofacitinib (n = 4, 5 mg twice a day) in conjunction with a stable dose of MTX. A second knee arthrocentesis was performed one month later to collect SF cells from the same joint.
SF cells isolation
The patient’s knee joint synovial fluid was extracted under aseptic conditions. Hyaluronidase (20426ES60, Yeasen) was added to the fresh SF sample obtained at a final concentration of 100 U/ml and incubated at 37 °C for 30 min. SF samples from each RA patient were obtained from the same joint longitudinal to avoid potential bias.
Cell suspension preparation
To prepare single-cell suspensions for sequencing needs, the fresh SF samples were centrifuged at 800 × g for 10 min, the supernatant was collected and stored at −80 °C, while the cell pellets were suspended by 1 ml red blood cell lysis buffer (R1010, Solarbio) to lyse the red blood cells and incubated on ice for 6 min, followed by centrifugation at 800 × g for 5 min. Next, 800 μl of pre-chilled PBS (BL302A, Biosharp) and 300 μl of pre-chilled debris removal solution (130-109-398, Miltenyi; reagent for cell debris removal through density gradient centrifugation) were added to suspend the cell pellets. After gently adding additional 800ul PBS and centrifuging at 800 × g for 5 min, the liquid will be divided into three phases. The top two phases were discarded, and 2 ml PBS were added to wash the lower phase of pellets once, centrifugation was performed at 800 × g for 5 min to remove contaminants and obtain pure cells. Finally, we collected a 20 μl aliquot for cell counting after discarding the supernatant and resuspended it by adding 500 μl PBS. All centrifugation procedures were carried out at 4 °C. Finally, high-quality single-cell suspensions were prepared for scRNA-seq.
Single cell library preparation and sequencing
The cell suspension was placed into chromium microfluidic chips, and a 10 × Chromium Controller (10× Genomics) was used to barcode the samples. Next, using components from a Chromium Single Cell 3’ (v3) reagent kit (10× Genomics), sequencing libraries were created using RNA from the barcoded cells after reverse transcription, in accordance with the manufacturer’s instructions. Sequencing was carried out using the Illumina NovaSeq 6000 platform in accordance with the manufacturer’s instructions. Single cell library preparation and subsequent next generation sequencing was performed by Novogene Co., Ltd. All the scRNA-seq data has passed the quality control.
Data processing and analysis
The scRNA-seq data was analyzed using our previously established workflow15,16,17,18,19,20,21,22,23. Briefly, raw FASTQ files generated by 10× Genomics were aligned and quantified using Cell Ranger (v4.3.0) against the GRCh38 human reference genome (v6.1.2). The Read10X function of the Seurat package was used to read the output of Cell Ranger. Doublets were identified and removed using the scrublet tool in Python, with an expected doublet rate of 0.1. Next, to facilitate downstream analyses, cells were distinctly labeled using the RenameCells function and integrated into a single aggregate object using the Merge function. To ensure data quality, stringent filtering parameters were applied to weed out any empty oil beads, mortality, and doublet cells, excluding cells with fewer than 200 or more than 4,000 detected genes, or with mitochondrial content exceeding 15%. Global scaling normalization was performed using the ‘LogNormalize’ method with a scaling factor of 10,000 to equalize overall gene expression across cells. The FindVariableFeatures function identified the top 2,000 highly variable genes, which were used for dimensionality reduction via principal component analysis (PCA). The first 30 principal components were selected, and the ScaleData function was applied to scale the expression of all genes, followed by regression of mitochondrial content to remove confounding variation. Sample batch effect correction was performed using the RunFastMNN function24, which is based on a multi-canonical correlation analysis algorithm. For clustering, the FindNeighbors and FindClusters functions with the built-in Louvain method were applied. Clusters were visualized using uniform manifold approximation and projection (UMAP). Cluster-specific marker genes were identified using the non-parametric Wilcoxon rank-sum test with Bonferroni correction (adjusted p value < 0.05) implemented in the FindAllMarkers function (min.pct = 0.25, logfc.threshold = 0.25). Cell populations were annotated based on canonical marker genes from published literature. Due to the inherent instability of neutrophils during processing and their predominant origin from a small number of samples, this cell type was excluded from subsequent analyses to mitigate technical bias. The proportion of each cluster was estimated by dividing the count of cells within each cluster by the total cell count in the respective sample. These ratios were statistically compared across treatment groups to assess significant differences in cell composition. The detailed workflow for data processing, including the steps for transferring data and generating figures and analyses presented in this study, is available on our GitHub repository at (https://github.com/Xiaxy-XuLab/RA-SF-data_discriptor).
Data Records
The dataset is available at the National Center for Biotechnology Information (NCBl) Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo) under accession number GSE29611725. To facilitate the reproduction of single-cell transcriptomic profiling, RDS (a file format to save Seurat objects) files for the RA SF scRNA-seq samples are included in the same GEO accession. The raw sequence data generated in this study have been deposited in the Genome Sequence Archive at National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA011646) and are available under accession number HRA01164626.
Technical Validation
Following SF cell isolation, cell suspension preparation, and single-cell library construction, scRNA-seq was conducted (Fig. 1). The raw scRNA-seq metrics for each sample are summarized in Supplementary Table 3. The total number of reads per sample exceeded 342 million. The Q20 base ranged from 94.34% to 97.22%, Q30 from 89.28% to 93.84%, and the GC content centered around 45%. Cell-based quality control metrics (Supplementary Table 4) showed that each sample contained more than 2,000 captured cells, with mean reads per cell ranging from 31,819 to 192,851 and median UMI counts per cell ranging from 2,420 to 10,387. The number of genes detected per sample varied from 11,318 to 23,562, with the median genes per cell ranging from 1,259 to 2,875. Sequencing saturation levels ranged from 60% to 90%, and potential doublets per sample ranged from 40 to 811 (expected doublet rate: 0.1). Mitochondrial RNA (mtRNA) levels were consistently low, reflecting high cell quality. After rigorous quality control, we obtained 100,387 high-quality cells across 18 samples, with cell counts per sample detailed in Supplementary Table 4. In summary, we generated high-quality single-cell transcriptomic data from nine matched pairs of synovial fluid samples collected from RA patients before and after treatment with adalimumab/tofacitinib.
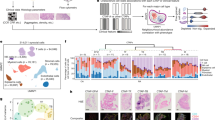
Schematic overview of experimental design. We collected SF samples from the knee joint of patients with RA who had not received b/tsDMARDs treatment at least in the last three months. RA patients were treated with adalimumab (40 mg every 2 weeks) or tofacitinib (5 mg twice a day) for 1 month after enrollment. After 1 month, patients with RA receive the second arthrocentesis of the knee to obtain the SF from the same joint. After the SF cells isolation, cell suspension preparation and single cell library preparation, a single-cell RNA sequencing was performed. RA, rheumatoid arthritis; ADA, adalimumab; TOF, tofacitinib.
To elucidate the cellular composition of SF from these RA patients, we employed the Seurat software package to stratify cells that passed quality control. A total of seven major cell clusters were identified and visualized using UMAP analysis (Fig. 2a). Cell clusters were annotated based on canonical marker genes derived from published literature27,28,29,30,31, as shown in a dot plot (Fig. 2b). Fibroblasts, a major component of ST samples in RA according to previous reports32,33, were present in lower proportions in SF and were primarily detected in one patient, while macrophages and T cells were the predominant cell types in SF (Fig. 3). Overall, we presented the cellular composition of each sample (Fig. 3a) and provided a statistical analysis of the cellular proportions based on the treatment status of samples (Fig. 3b).
Cell clustering and identification of scRNA-Seq samples. (a) Visualization of seven main clusters across 100,387 cells using uniform manifold approximation and projection (UMAP). (b) Dot plot illustrating the normalized expression level of cell type-specific marker genes across SF clusters. The dots’ size and color spectrum indicate positive percentage and average expression (log1p transformed) of particular markers genes in each cell type, respectively. (c) Visualization of cells based on the treatment status of samples by using UMAP. BT, before treatment; AT, after treatment.
Proportion analysis of cell clusters. (a) Quantification of relative proportions of distinct cell clusters in SF among patients. The horizontal coordinates depict cell proportions, the vertical coordinates depict patients. The left and right sides represent samples before and after treatment, respectively. (b) Proportional analysis. Proportions of distinct primary cell clusters within each group.
To investigate the molecular changes induced by adalimumab/tofacitinib treatment, we profiled the expression of established RA-related genes across cell types. Consistent with previous scRNA-seq-based findings in ST samples from RA32,34,35, IL6, IL1A, IL12A, and IL12B exhibited low positive expression, while JAK1 and JAK3 were expressed at higher levels compared to TNF (Fig. 4a). Expression of JAK1 and JAK3 was significantly elevated in RA-BT samples and decreased following treatment with adalimumab/tofacitinib (Fig. 4b). While the quantitative validation confirms the persistent low expression of JAK2/3 across most cellular subpopulations, tofacitinib selectively downregulated JAK1/JAK2/JAK3 in T cells and JAK2/JAK3 in macrophages, whereas adalimumab showed no such specificity (Fig. 4b).
Expression of disease-related genes in SF cells. (a) Violin Plot depicting the expression level of well-reported RA-related genes in the major cell types. (b) Dot plot illustrating the pairwise comparisons of expression levels of well-reported RA-related genes in different cell clusters. The p values were calculated using a two-sided Wilcoxon test. The dot size and color spectrum indicate p value (-log10 transformed) and fold change (log2 transformed) of gene expression, respectively.
Usage Notes
Our dataset comprises scRNA-seq data derived from SF samples of RA patients collected before and after treatment with tofacitinib/adalimumab. It provides a comprehensive profile of cellular composition, molecular signaling pathways, and functional characteristics. This dataset offers a valuable resource for addressing a range of research questions, including elucidating the mechanisms of action of tofacitinib and adalimumab, identifying novel therapeutic targets for RA, and exploring the pathogenesis of other inflammatory diseases. The raw sequence data generated in this study have been deposited in the GSA-human database, researchers can apply to the Data Access Committee (DAC) via the GSA platform. DAC approval for access to the dataset will be granted to all users that agree to the terms and conditions of the data access agreement. For detailed application procedures, please refer to the official guide (https://ngdc.cncb.ac.cn/gsa-human/document/GSA-Human_Request_Guide_for_Users_us.pdf), and data access requests are typically processed within two weeks.
Code availability
The bioinformatics analysis code is available on gitHub at (https://github.com/Xiaxy-XuLab/RA-SF-data_discriptor).
References
Smolen, J. S. et al. Rheumatoid arthritis. Nat Rev Dis Primers 4, 18001, https://doi.org/10.1038/nrdp.2018.1 (2018).
Smolen, J. S., Aletaha, D. & McInnes, I. B. Rheumatoid arthritis. Lancet 388, 2023–2038, https://doi.org/10.1016/S0140-6736(16)30173-8 (2016).
Boissier, M. C., Biton, J., Semerano, L., Decker, P. & Bessis, N. Origins of rheumatoid arthritis. Joint Bone Spine 87, 301–306, https://doi.org/10.1016/j.jbspin.2019.11.009 (2020).
Pappas, D. A. et al. Comparative effectiveness of first-line tumour necrosis factor inhibitor versus non-tumour necrosis factor inhibitor biologics and targeted synthetic agents in patients with rheumatoid arthritis: results from a large US registry study. Ann Rheum Dis 80, 96–102, https://doi.org/10.1136/annrheumdis-2020-217209 (2021).
Combe, B. et al. Filgotinib versus placebo or adalimumab in patients with rheumatoid arthritis and inadequate response to methotrexate: a phase III randomised clinical trial. Ann Rheum Dis 80, 848–858, https://doi.org/10.1136/annrheumdis-2020-219214 (2021).
Fitton, J., Melville, A. R., Emery, P., Nam, J. L. & Buch, M. H. Real-world single centre use of JAK inhibitors across the rheumatoid arthritis pathway. Rheumatology (Oxford) 60, 4048–4054, https://doi.org/10.1093/rheumatology/keaa858 (2021).
Taylor, P. C. et al. Baricitinib versus Placebo or Adalimumab in Rheumatoid Arthritis. N Engl J Med 376, 652–662, https://doi.org/10.1056/NEJMoa1608345 (2017).
Aletaha, D. & Smolen, J. S. Diagnosis and Management of Rheumatoid Arthritis: A Review. JAMA 320, 1360–1372, https://doi.org/10.1001/jama.2018.13103 (2018).
Smolen, J. S. et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann Rheum Dis 79, 685–699, https://doi.org/10.1136/annrheumdis-2019-216655 (2020).
Fraenkel, L. et al. American College of Rheumatology Guideline for the Treatment of Rheumatoid Arthritis. Arthritis Care Res (Hoboken) 73, 924–939, https://doi.org/10.1002/acr.24596 (2021).
Winthrop, K. L. & Cohen, S. B. Oral surveillance and JAK inhibitor safety: the theory of relativity. Nat Rev Rheumatol 18, 301–304, https://doi.org/10.1038/s41584-022-00767-7 (2022).
Liao, H. et al. Use of mass spectrometry to identify protein biomarkers of disease severity in the synovial fluid and serum of patients with rheumatoid arthritis. Arthritis Rheum 50, 3792–3803, https://doi.org/10.1002/art.20720 (2004).
Bhattacharjee, M. et al. Synovial fluid proteome in rheumatoid arthritis. Clin Proteomics 13, 12, https://doi.org/10.1186/s12014-016-9113-1 (2016).
Aletaha, D. et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum 62, 2569–2581, https://doi.org/10.1002/art.27584 (2010).
Luo, H. et al. Characterizing dedifferentiation of thyroid cancer by integrated analysis. Sci Adv 7, https://doi.org/10.1126/sciadv.abf3657 (2021).
Luo, H. et al. Pan-cancer single-cell analysis reveals the heterogeneity and plasticity of cancer-associated fibroblasts in the tumor microenvironment. Nature communications 13, 6619, https://doi.org/10.1038/s41467-022-34395-2 (2022).
Cao, M. et al. Characterization of immature ovarian teratomas through single-cell transcriptome. Frontiers in immunology 14, 1131814, https://doi.org/10.3389/fimmu.2023.1131814 (2023).
Yin, D. et al. Novel insights into HBV‐hepatocellular carcinoma at single‐cell sequencing. MedComm–Oncology 2, e60 (2023).
Mao, W. et al. Pan-cancer single-cell landscape of drug-metabolizing enzyme genes. Pharmacogenetics and genomics 34, 217–225, https://doi.org/10.1097/fpc.0000000000000538 (2024).
Zhou, T. et al. The role of long non‐coding RNA Maternally Expressed Gene 3 in cancer‐associated fibroblasts at single cell pan‐cancer level. Interdisciplinary Medicine 2, e20240018 (2024).
Cao, M. et al. Single-cell transcriptomic analysis reveals gut microbiota-immunotherapy synergy through modulating tumor microenvironment. Signal Transduct Target Ther 10, 140, https://doi.org/10.1038/s41392-025-02226-7 (2025).
Wang, Y. et al. Single-Cell Transcriptome Analysis Reveals Interaction between CCL19+ Inflammatory Keratinocytes and CCR7+ Dendritic Cells and B Cells in Pemphigus. J Invest Dermatol 144, 2301–2305 e2308, https://doi.org/10.1016/j.jid.2024.03.016 (2024).
Li, P. H. et al. Contribution of crosstalk of mesothelial and tumoral epithelial cells in pleural metastasis of lung cancer. Transl Lung Cancer Res 13, 965–985, https://doi.org/10.21037/tlcr-24-118 (2024).
Haghverdi, L., Lun, A. T. L., Morgan, M. D. & Marioni, J. C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 36, 421–427, https://doi.org/10.1038/nbt.4091 (2018).
He, C. et al. A single-cell RNA-seq dataset of synovial fluid from rheumatoid arthritis treated with TNF-α/JAK inhibitor. Gene Expression Omnibus, https://identifiers.org/geo/GSE296117 (2025).
National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences. https://ngdc.cncb.ac.cn/gsa-human/browse/HRA011646 (2025).
Shaul, M. E. & Fridlender, Z. G. Tumour-associated neutrophils in patients with cancer. Nature reviews. Clinical oncology 16, 601–620, https://doi.org/10.1038/s41571-019-0222-4 (2019).
Zilionis, R. et al. Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity 50, 1317–1334.e1310, https://doi.org/10.1016/j.immuni.2019.03.009 (2019).
Zhang, L. et al. Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell 181, 442–459 e429, https://doi.org/10.1016/j.cell.2020.03.048 (2020).
Schelker, M. et al. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nature communications 8, 2032, https://doi.org/10.1038/s41467-017-02289-3 (2017).
Xia, X. et al. Single cell immunoprofile of synovial fluid in rheumatoid arthritis with TNF/JAK inhibitor treatment. Nat Commun 16, 2152, https://doi.org/10.1038/s41467-025-57361-0 (2025).
Zhang, F. et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol 20, 928–942, https://doi.org/10.1038/s41590-019-0378-1 (2019).
Weyand, C. M. & Goronzy, J. J. The immunology of rheumatoid arthritis. Nat Immunol 22, 10–18, https://doi.org/10.1038/s41590-020-00816-x (2021).
Walker, J. G. et al. Characterisation of a dendritic cell subset in synovial tissue which strongly expresses Jak/STAT transcription factors from patients with rheumatoid arthritis. Ann Rheum Dis 66, 992–999, https://doi.org/10.1136/ard.2006.060822 (2007).
Rossi, J. F., Lu, Z. Y., Jourdan, M. & Klein, B. Interleukin-6 as a therapeutic target. Clin Cancer Res 21, 1248–1257, https://doi.org/10.1158/1078-0432.CCR-14-2291 (2015).
Acknowledgements
This study was supported by the National Key R&D Program of China (No. 2021YFA1301203(H.X.)), National Natural Science Foundation of China (81973408(H.X.)), the China Postdoctoral Science Foundation (2022M712274(X.Y.X.)), Sichuan Science and Technology Program, 2023NSFSC1704(X.Y.X.), 2024YFFK0062(Q.B.X), 2025ZNSFSC1745(C.J.H.), 2025ZNSFSC0638(G.Y.), 2025ZNSFSC0046(H.X.)), 1.3.5 Project for Disciplines of Excellence, West China Hospital of Sichuan University (ZYYC23025 (H.X)).
Author information
Authors and Affiliations
Contributions
G.Y., Q.B.X. and H.X. designed and supervised the project. Z.X.R.,X.Y.X., M.X.Q. and Y.S. analyzed the data. X.Y.X., F. Z., H.X. and Q.B.X interpreted the data. C.J.H., Z.N.X., and and G.Y. collected the clinical samples and information. C.J.H. and X.Y.X. draw the diagrams. C.J.H., Z.X.R., X.Y.X. and H.X. wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
He, C., Ren, Z., Xia, X. et al. A single-cell RNA-seq dataset of synovial fluid from rheumatoid arthritis treated with TNF-α/JAK inhibitor. Sci Data 12, 977 (2025). https://doi.org/10.1038/s41597-025-05339-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-025-05339-4