Abstract
The English green aphid, Sitobion avenae, a major pest of wheat, exhibits classical wing dimorphism. To support research and data sharing on the molecular basis of this trait, we generated full-length transcriptomes from three different developmental stages of winged and wingless morphs using PacBio SMRT and Illumina HiSeq sequencing platforms. The dataset comprises 2,309,013 circular consensus sequences (CCSs), with 85.29% identified as full-length non-chimeric reads (FLNC) reads after filtering. Approximately 282 Gb of PacBio subreads were obtained, with a total of 125,495,799 reads. Functional annotation was performed for 43,219 transcripts (44.3%). Across the developmental stages, differential expression analyses revealed numerous genes with varied expression patterns, with 71 genes identified as potential regulators of wing polymorphism. These candidates are associated with biological processes such as wing development, hormone biosynthesis, energy metabolism, and cell death pathways. This dataset provides a comprehensive molecular resource for investigating the transcriptional basis of wing polyphenism in aphids and may offer insights applicable to other insect systems.
Similar content being viewed by others
Background & Summary
Aphids (Hemiptera: Aphididae) include numerous species, approximately 450 of which are recognized as serious agricultural pests worldwide, causing significant economic losses due to direct feeding and virus transmission1,2. The grain aphid, Sitobion avenae, is one of the major wheat pests across temperate regions, where it extracts phloem sap, leading to stunted growth, premature aging of tissues, or even deformity3. Additionally, the sticky honeydew excreted by aphids facilitates the growth of sooty mold, which disrupts photosynthesis and further degrades wheat quality. Due to the immense losses caused by S. avenae, it has been designated as a first-class pest in China by the Chinese Ministry of Agriculture and Rural Affairs for many consecutive years.
The migration of insects is an adaptive trait that has evolved over time to cope with environmental changes4,5. In S. avenae, this adaptability is exemplified by its wing dimorphism—a critical survival strategy that significantly amplifies its agricultural impact. As a wing polymorphic species, S. avenae serves as an ideal model for studying life history adaptation mechanisms. Its predominantly parthenogenetic life cycle allows the same genotype to produce two distinct morphs: winged individuals capable of long-distance migration, facilitating rapid dispersal across wheat fields, and wingless individuals that maintain high-density populations through localized reproduction6. Environmental cues including temperature7, population density8, maternal effects9, and hormonal signaling (e.g., juvenile hormone, molting hormone, and insulin), jointly regulate this developmental plasticity. However, despite extensive studies on the environmental and physiological determinants of wing dimorphism, the molecular mechanisms that translate environmental signals into wing morph outcomes remain poorly understood. To address this gap, our study employs high-throughput transcriptomic analysis to identify key regulatory genes and pathways involved in wing differentiation in S. avenae, which may contribute to a deeper understanding of the genetic architecture underlying aphid polyphenism.
Recent advances in sequencing technologies have revealed critical genes and signaling pathways that regulate aphid wing polymorphism. These pathways can be categorized into two major mechanisms. The first involves hormone signaling pathways, such as the molting hormone signaling pathway that mediates intergenerational wing polymorphism in Acyrthosiphon pisum10, and miR-9b, which negatively regulates ABCG4 expression in Aphis citricidus, ABCG4 influences insulin signaling pathway activity by modulating IRP3 expression, thereby regulating wing development11. The second mechanism involves cell autophagy or apoptosis pathways. For instance, TOR regulates wing primordium autophagy and the proportion of winged aphids in A. pisum by inhibiting REPTOR2 transcription12, while the Wnt signaling pathway modulates wing polymorphism in pea aphids by inhibiting cell apoptosis13. While significant progress has been made in understanding the genes and the hormonal regulation of wing polyphenism in other insect species, the precise molecular mechanisms underlying wing dimorphism in S. avenae remain poorly understood.
Transcriptome serves as a crucial link between genomic and proteomic data, providing insights into gene function and regulation. Second-generation sequencing technologies, such as RNA sequencing (RNA-Seq) via the Illumina platform, have revolutionized research on insect pests, particularly in pest management and control strategies14,15. This approach fragments DNA into short sequences (400–500 bp) for high-throughput analysis16. However, assembling full-length transcripts from short reads often results in incomplete data, necessitating the use of complementary technologies. Third-generation sequencing technologies, such as PacBio’s Single Molecule Real-Time (SMRT) sequencing, overcome the limitations of short-read sequences by reducing information loss and base mismatches, offering a more comprehensive view of transcriptomic complexity17. Despite its advantages, SMRT sequencing faces challenges, including high error rates, which can be mitigated by increasing sequencing coverage or integrating data with second-generation sequencing18,19.
In this study, we conducted a temporal transcriptomic analysis to investigate wing dimorphism in S. avenae across three developmental stages, including pseudo embryo, 1st and 2nd instar nymphs, from both winged and wingless morphs. Using a combination of SMRT and Illumina sequencing platforms, we generated full-length transcript annotations and characterized long noncoding RNAs (lncRNAs), transcription factors (TFs), and alternative splicing events. The schematic overview of the study design and workflow is shown in Fig. 1. The transcriptomic resources generated in this study offer insights into the gene networks underling wing differentiation, which may aid in further understanding wing dimorphism in aphids and other insects.
Schematic workflow of full-length transcriptome analysis for wing dimorphism in Sitobion avenae. The analysis begins with the collection of winged and wingless nymph samples, followed by Iso-Seq and RNA-Seq. The obtained data are then corrected and aligned to the reference genome. Subsequent analyses include database annotations, structural analysis, and differential expression analysis to identify genes associated with wing dimorphism. Pe: pseudo embryo; N1-N2: first to second instar nymphs; NR (Non-redundant protein database), NT (Non-redundant nucleotide database), GO (Gene Ontology), Pfam (Protein families database), KOG (euKaryotic Ortholog Groups), KEGG (Kyoto Encyclopedia of Genes and Genomes), Swiss-Prot (manually annotated and reviewed protein sequence database), AS (Alternative splicing), APA (Alternative Polyadenylation), LncRNA (Long non-coding RNA), DEG (Differential Expressed Genes).
Methods
Samples selection and preparation
Sitobion avenae (F.) specimens were collected from common wheat (T. aestivum) fields in Langfang, Hebei Province, China (39°30′42′′N, 116°36′7′′E) in 2018. The aphids were transferred to the laboratory and reared on 15 cm-tall wheat seedlings under controlled conditions of 20 °C, 60% RH, and 16:8 h L:D photoperiod. All experiments utilized clonal female progeny from the third generation (G3). To investigate morph-specific transcriptomes, six representative sample types were collected from three developmental stages of winged and wingless S. avenae. Winged morphs were induced by crowding and alarm pheromone exposure, wherein G3 nymphs were reared in 9 cm Petri dishes and treated with 5,000 ng of EβF (1000 ng/μL applied at five time points per day) for five consecutive days20. From these groups, winged pseudo-embryos (W.Pe) were obtained by dissecting G4 adults from the first group, while first-instar (W.N1) and second-instar (W.N2) winged nymphs were collected based on molting stages from the offspring of the other two groups. In parallel, wingless morphs were produced by rearing genetically identical clonal nymphs under identical conditions with water treatment instead of EβF. Corresponding wingless pseudo-embryos (WL.Pe) were dissected from G4 wingless adults, and wingless first-instar (WL.N1) and second-instar (WL.N2) nymphs were collected from their progeny. All samples were immediately frozen in liquid nitrogen and stored at –80 °C for RNA extraction and sequencing.
RNA extraction
Total RNA was extracted from each sample using TRIzol reagent and the RNeasy Plus Mini Kit (Qiagen, Valencia, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit and analyzed with the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
PacBio library construction and sequencing
For the PacBio sequencing library construction, an equal amount of high-quality RNA from the fourth replicate of each sample, after being pooled with the corresponding replicates from the same developmental stage and wing type, was processed for sequencing at Novogene (Beijing, China). Eukaryotic mRNA with a poly-A tail was reverse-transcribed into cDNA using Oligo(dT) primers. The full-length cDNA was then amplified using PCR and purified with PB magnetic beads to remove cDNA fragments shorter than 1 kb, and then ligated to SMRT dumbbell adapters. Unconnected fragments were digested by exonuclease and purified again with PB magnetic beads to create the sequencing library. The final library was quantified using a Qubit 3.0 fluorometer (Thermo Fisher Scientific) and assessed for fragment size distribution with an Agilent 2100 Bioanalyzer (Agilent Technologies). Raw PacBio subreads were processed using the SMRT Link software suite (v11.0.0.146107, Pacific Biosciences). Circular consensus sequences (CCS) were generated using the CCS algorithm21. Full-length non-chimeric (FLNC) reads were then identified and clustered using the Iso-Seq pipeline22. Sequences shorter than 50 bp were excluded from downstream analyses.
Illumina Library construction and sequencing
mRNA from three distinct developmental stages of both winged and wingless aphids of S. avenae was isolated using Oligo(dT) magnetic beads. RNA purity and integrity were assessed using the NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and the Bioanalyzer 2100 system. RNA contamination was evaluated through 1.5% agarose gel electrophoresis. mRNA purification was conducted according to the manufacturer’s protocol, and library construction was performed using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA). Library quality and size were evaluated with the Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and the Agilent 2100 system. Sequencing was subsequently conducted on the Novaseq 6000 platform (Illumina, CA, USA) at Novogene (Beijing, China).
PacBio Iso-Seq data processing
Full-length transcriptomes of S. avenae were generated using the PacBio Sequel platform, based on total RNA extracted from six distinct developmental stages. Each developmental stage was sequenced independently, resulting in six separate Iso-Seq datasets that enabled stage-specific resolution of transcript isoforms (Table 1). A total of 125,495,799 subreads were generated, producing approximately 281.92 Gb of raw sequencing data. The average subread length ranged from 1,658 to 2,789 bp, with an overall N50 of 3,217 bp. The sequencing yielded 2,309,013 CCSs, of which 86.40% were identified as full-length reads. After removal of chimeric sequences, 85.29% of the reads were retained as high-confidence full-length non-chimeric (FLNC). The average FLNC read lengths ranged from 2,013 bp to 3,128 bp across developmental stages. Error correction was performed using Illumina short-read data, and high-quality corrected consensus isoforms were generated using LoRDEC. For each sample, both the total nucleotide counts and N50/N90 values showed minimal differences before and after correction (Table 2).
Functional annotation
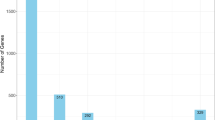
To validate the accuracy of Iso-Seq reads, we used the GMAP (Genomic Mapping and Alignment Program) software23 to map the corrected high-quality consensus sequences to the reference genome of the Asian wheat aphid, Sitobion miscanthi (http://gigadb.org/dataset/100635)24. Transcripts with ≥95% identity and ≥99% coverage were merged to remove redundancy. Isoforms were categorized into three groups: unmapped, multiple mapped, and uniquely mapped (Table 3). Unmapped transcript sequences were functionally annotated by querying seven databases: NCBI non-redundant proteins (NR)25, NCBI non-redundant nucleotide sequences (NT), Gene Ontology (GO)26, Kyoto Encyclopedia of Genes and Genomes (KEGG)27, euKaryotic Ortholog Groups (KOG)28, Protein Family (Pfam)29, and manually annotated and reviewed protein sequences (Swiss-Prot)30 databases. KEGG pathway enrichment analysis was conducted using the KOBAS software (http://kobas.cbi.pku.edu.cn/home.do). A total of 43,219 unigenes were annotated, achieving a 99.59% annotation rate across at least one database (Fig. 2).
Functional annotation of transcripts across seven databases: NR (Non-redundant protein database), NT (Non-redundant nucleotide database), Swiss-Prot (A manually annotated and reviewed protein sequence database), GO (Gene Ontology), Pfam (Protein families database), KEGG (Kyoto Encyclopedia of Genes and Genomes), and KOG (euKaryotic Ortholog Groups).
lncRNAs and transcription factors
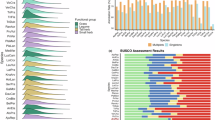
Long non-coding RNAs (lncRNAs) were predicted using CNCI, CPC, Pfam, and Plek tool, identifying 6,058, 752, 4,091, and 7,645 lncRNAs, respectively. A total of 596 lncRNAs were consistently predicted by all four methods (Fig. 3). Additionally, 1,398 transcription factors (TFs) were identified, with enriched TF families including zf-C2H2 (306), ZBTB (169), HMG (160), MYB (120), and Homeobox (65) (Fig. 4).
Candidate long non-coding RNAs (lncRNAs) identified using a combination of four tools: CNCI (Coding-Non-Coding Index), CPC (Coding Potential Calculator), Pfam (Protein families database, used for excluding protein-coding genes), and Plek (a tool for predicting long non-coding RNAs and messenger RNAs based on k-mer content and other features).
Differentially expressed genes
Gene expression data from PacBio SMRT and Illumina sequencing platforms were compared across developmental stages to identify differentially expressed genes (DEGs). Differential expression analysis was conducted using the edgeR package31. Data were normalized using the trimmed mean of M-values (TMM) method to account for compositional biases in RNA-seq data, enabling accurate comparisons across samples. Two types of comparisons were performed: (i) between morphs, comparing gene expressions between winged and wingless aphids at the same developmental stage, and (ii) within morphs, comparing gene expressions across developmental stages of the same morphotype. In each pairwise comparison, “group1” corresponds to samples from the first condition (e.g., winged N1), and “group2” to the second condition (e.g., wingless N1 or winged N2). DEGs were identified using a threshold of |log2 (group1/group2)| ≥1, with FDR <0.001 and P < 0.005. Gene expression levels were subsequently quantified as FPKM values to compare expression patterns across the six developmental stages. To visualize the gene expression patterns, clustering methods including H-clustering (hierarchical clustering), K-means clustering, and SOM (self-organizing maps) were applied. The resulting clusters were visualized as heatmaps (Fig. 5).
Cluster analysis of differentially expressed genes (DEGs) across developmental stages of S. avenae. This heatmap presents hierarchical clustering of FPKM values. The log10(FPKM + 1) values were normalized (scaled) and subjected to clustering analysis. The gradient from red to blue represents a decrease in log10(FPKM + 1) values, representing a transition from high to low expression. W.Pe: winged pseudo embryo; W.N1: winged 1st instar nymphs; W.N2: winged 2nd instar nymphs; WL.Pe: wingless pseudo embryo; WL.N1: wingless 1st instar nymphs; WL.N2: wingless 2nd instar nymphs.
Functional categorization of candidate genes
To facilitate downstream exploration of wing polymorphism in S. avenae, KEGG pathway enrichment analysis was conducted using a subset of 71 genes selected from differential expression results. These genes are involved in diverse biological processes, including wing development, hormone synthesis, and energy metabolism. Enriched pathways identified from comparisons among different morphs and developmental stages included ECM-receptor interaction, ABC transporters, Carbon metabolism, Fatty acid metabolism, and insect hormone biosynthesis. A full list of enriched pathways is provided in Table S1. Gene expression levels across samples were visualized using a heatmap generated with Tbtools (Version 2.210)32 (Fig. 6).
Expression profile of differentially expressed genes (DEGs) across developmental stages of S. avenae. Each data block represents the log10 (FPKM + 1) values for the corresponding samples. W.Pe: winged pseudo embryo; W.N1: winged 1st instar nymphs; W.N2: winged 2nd instar nymphs; WL.Pe: wingless pseudo embryo; WL.N1: wingless 1st instar nymphs; WL.N2: wingless 2nd instar nymphs.
Data Records
All Transcriptome sequencing data generated in this study have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession numbers SRP546082 and SRP57534833,34. The dataset includes raw reads obtained from both the PacBio SMRT and Illumina platforms, covering three developmental stages (pseudo-embryo, 1st instar, and 2nd instar nymphs) of S. avenae in both winged and wingless morphs. Accompanying metadata, including experimental conditions, library preparation protocols, and sample annotations, are provided alongside the sequencing data to facilitate reuse and reproducibility. In addition to the raw sequencing data, the polished consensus transcript sequences, corresponding functional annotation files, and identified long non-coding RNA (lncRNA) derived from PacBio Iso-Seq data have been deposited in Figshare35. These processed files are publicly accessible and support downstream analyses and comparative transcriptomic studies.
Technical Validation
To analyze the overall mRNA expression pattern in S. avenae, full-length transcriptomic sequencing was performed across six developmental stages. Transcriptome completeness was assessed using BUSCO v5.6.1 with the arthropoda_odb10 dataset (1,013 BUSCOs), yielding a completeness score of 65.1%, comprising 41.5% single-copy, 23.6% duplicated, 3.2% fragmented, and 31.7% missing BUSCOs. These values are within the range reported for long-read transcriptome assemblies in non-model insects. To evaluate sequencing performance, short-read Illumina data were aligned to the reference genome using HISAT236, resulting in mapping rate exceeding 90% across all samples. Mapping rates were calculated as the percentage of reads aligning to the reference genome, provides an estimate of the sequencing accuracy and alignment efficiency. Sequencing quality metrics showed average Q20 and Q30 scores of 99.5% and 98.9%, respectively. Identity and coverage metrics were used to evaluate transcript quality and completeness. Identity refers to the sequence similarity between a transcript and its reference counterpart, while coverage represents the proportion of the reference gene sequence covered by the transcript. Transcripts with both coverage and identity greater than 75% were classified as high-quality, demonstrating good homology and sequencing reliability (Fig. 7). All reported metrics were used to assess the accuracy and integrity of the transcriptome dataset and to support its suitability for subsequent analyses.
Code availability
All software used in this study, including their versions and parameters, are described in the method section. For tools where parameters were not explicitly provided, default settings were applied. Data visualization plots were generated using GraphPad V.8.02.
References
Wenger, J. A. et al. Michel A. Whole genome 384 sequence of the soybean aphid, Aphis glycines. Insect. Biochem. Mol. Biol. 123, 102917, https://doi.org/10.1016/j.ibmb.2017.01.005 (2020).
Sorensen, J. T. Chapter 8 - Aphids. In Encyclopedia of Insects (Second Edition) eds. V. H. Resh & R. T. Cardé, 27-31. (San Diego: Academic Press, 2009).
Zhang, G. Aphids in Agriculture and Forestry of Northwest China 1st ed. (Beijing: China Environmental Science, (1999).
Roff, D. A. The evolution of wing dimorphism in insects. Evolution. 40, 1009–20, https://doi.org/10.1111/j.1558-5646.1986.tb00568.x (1986).
Roff, D. A. & Fairbairn, D. J. Wing dimorphisms and the evolution of migratory polymorphisms among the insect. Am. Zool. 31, 243–51, https://doi.org/10.1093/icb/31.1.243 (1991).
Harrison, R. G. Dispersal polymorphisms in insects. Annu. Rev. Ecol. Syst. 11, 95–118, https://doi.org/10.1146/annurev.es.11.110180.000523 (1980).
Parish, W. E. G. & Bale, J. S. Effects of short term exposure to low temperature on wing development in the grain aphid Sitobion avenae (F.) (Hem., Aphididae). J. Appl. Entomol. 109, 175–181, https://doi.org/10.1111/j.1439-0418.1990.tb00034.x (1990).
Dixon, A.F.G. Aphid Ecology 2nd Ed (Chapman and Hall, 1998).
Brisson, J. A. Aphid wing dimorphisms: linking environmental and genetic control of trait variation. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 365, 1540, https://doi.org/10.1098/rstb.2009.0255 (2010).
Vellichirammal N.N., Gupta P., Hall T.A. & Brisson J.A. Ecdysone signaling underlies the pea aphid transgenerational wing polyphenism. Proc. Natl. Acad. Sci. USA 114, 6, https://doi.org/10.1073/pnas.1617640114 (2017).
Shang F. et al. The miR-9b microRNA mediates dimorphism and development of wing in aphids. Proc. Natl. Acad. Sci. USA 117, 15, https://doi.org/10.1073/pnas.1919204117 (2020).
Yuan, E. et al. A novel gene REPTOR2 activates the autophagic degradation of wing disc in pea aphid. Elife. 12, e83023, https://doi.org/10.7554/eLife.83023 (2023).
Zhou, P. et al. The Wnt pathway regulates wing morph determination in Acyrthosiphon pisum. Insect Biochem Mol Biol. 161, 104003, https://doi.org/10.1016/j.ibmb.2023.104003 (2023).
Bao, Y. Y. et al. The genome- and transcriptome-wide analysis of innate immunity in the brown plan-thopper, Nilaparvata lugens. BMC Genomics 14, 160, https://doi.org/10.1186/1471-2164-14-160 (2013).
He, S. et al. Comparative analysis of transcriptomes among Bombyx mori strains and sexes reveals the genes regulating melanic morph and the related phenotypes. PLoS One 11, e0155061, https://doi.org/10.1371/journal.pone.0155061 (2016).
Heather, J. M. & Chain, B. The sequence of sequencers: The history of sequencing DNA. Genomics. 107, 1–8, https://doi.org/10.1016/j.ygeno.2015.11.003 (2016).
Rhoads, A. & Au, K. F. PacBio sequencing and its applications. Genom. Proteom. Bioinf. 13, 278–289, https://doi.org/10.1016/j.gpb.2015.08.002 (2015).
Au, K. F., Underwood, J. G., Lee, L. & Wong, W. H. Improving PacBio long read accuracy by short read alignment. PLoS One 7, e46679, https://doi.org/10.1371/journal.pone.0046679 (2012).
Xu, Z. et al. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant. J. 82, 951–961, https://doi.org/10.1111/tpj.12865 (2015).
Li, X. et al. Temporal analysis of microRNAs associated with wing development in the English grain aphid, Sitobion avenae (F.) (Homoptera: Aphidiae). Insect Biochem. Mol. Biol. 142, 103579, https://doi.org/10.1016/j.ibmb.2021.103579 (2022).
Wenger, A. M. et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 37, 1155–1162, https://doi.org/10.1038/s41587-019-0217-9 (2019).
Sharon, D. et al. A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 31, 1009–1014, https://doi.org/10.1038/nbt.2705 (2013).
Wu, T. D. & Watanabe, C. K. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859, https://doi.org/10.1093/bioinformatics/bti310 (2005).
Jiang, X. et al. A chromosome-level draft genome of the grain aphid Sitobion miscanthi. Gigascience. 8(8), giz101, https://doi.org/10.1093/gigascience/giz101 (2019). 1.
Li, W., Jaroszewski, L. & Godzik, A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 18, 77–82, https://doi.org/10.1093/bioinformatics/18.1.77 (2002).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29, https://doi.org/10.1016/10.1038/75556 (2000).
Kanehisa, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, 277D–280D, https://doi.org/10.1093/nar/gkh063 (2004).
Koonin, E. V. et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 5, R7, https://doi.org/10.1186/gb-2004-5-2-r7 (2004).
Punta, M. et al. The Pfam protein families database. Nucleic Acids Res 40, D290–D301, https://doi.org/10.1093/nar/gkr1065 (2011).
Apweiler, R. UniProt: the universal protein knowledgebase. Nucleic Acids Res 32, 115D–119D, https://doi.org/10.1093/nar/gky092 (2004).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–40, https://doi.org/10.1093/bioinformatics/btp616 (2010).
Chen, C. et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202, https://doi.org/10.1016/j.molp.2020.06.009 (2020).
NCBI Sequence Read Archive http://identifiers.org/ncbi/insdc.sra:SRP546082 (2025).
NCBI Sequence Read Archive http://identifiers.org/ncbi/insdc.sra:SRP575348 (2025).
Yan, Y. et al. Comprehensive full-Length transcriptomic resource for wing dimorphism in English green aphid Sitobion avenae. Figshare https://doi.org/10.6084/m9.figshare.28822115 (2025).
Kim, M. D., Langmead, B. & Salzberg, S. L. Hisat: a fast spliced aligner with low memory requirements. Nat. Methods. 12, 357–360, https://doi.org/10.1038/nmeth.3317 (2015).
Acknowledgements
This research was supported by the National Natural Science Foundation of China (Award number: 32372547), and the earmarked fund for CARS (CARS-03). Authors would like to thank anonymous reviewers for their constructive criticisms and comments to improve this manuscript.
Author information
Authors and Affiliations
Contributions
X.R.L. conceived and designed the experiments. Y.Y., Q.T.L., G.H.W., Y.X.G., L.Y. W. and C.Z. collected and processed the tissue samples. Y.Y. carried out the experiments, analyzed the data and drafted the manuscript. Y.H.Z. and X.Z. contributed analysis tools and evaluated the results. X.R.L. and X.G.Z. provided critical analyses, and revised, wrote portions, and finalized the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41597_2025_5370_MOESM1_ESM.xlsx
Table S1. Genes Potentially Involved in Regulating Wing Polymorphism Across the Developmental Stages of Sitobion avenae.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yan, Y., Lv, Q., Wei, G. et al. Comprehensive full-Length transcriptomic resource for wing dimorphism in English green aphid, Sitobion avenae. Sci Data 12, 1034 (2025). https://doi.org/10.1038/s41597-025-05370-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05370-5