Abstract
The stag beetle (Coleoptera: Lucanidae) represents a captivating and evolutionarily significant group, regarded as one of the most basal lineages within the superfamily Scarabaeoidea. Despite their importance for studying beetle evolution and ecology, genomic resources for this family remain scarce. Here, we report a chromosome-level genome assembly of Odontolabis cuvera, generated by integrating PacBio HiFi, Illumina, and Hi-C data. The genome assembly spans 908.07 Mb, comprising 66 scaffolds (scaffold N50: 65.36 Mb) and 147 contigs (contig N50: 16.39 Mb). A total of 99.58% (904.22 Mb) of the assembly was anchored to 14 chromosomes. BUSCO analysis (insecta_odb10 dataset, n = 1,367) demonstrated high completeness, with 99.1% of conserved insect orthologs identified (98.3% single-copy, 0.8% duplicated). Repetitive elements accounted for 53.00% (281.28 Mb) of the genome, and a total of 18,332 protein-coding genes were annotated. This high-contiguity genome provides a critical foundation for uncovering the evolutionary mechanisms and ecological adaptations unique to Lucanidae.
Similar content being viewed by others
Background & Summary
Stag beetles (family Lucanidae) belong to the superfamily Scarabaeoidea within the order Coleoptera, comprising approximately 1,500 species distributed globally1. Male stag beetles are renowned for their enlarged mandibles, which they use in combative displays to secure preferred mating sites and food competition2. Owing to their striking morphology and complex behavior, many lucanid species have become model organisms for studies on behavioral ecology and functional morphology3. Their impressive mandibles also contribute to their popularity as exotic pets and valuable items in private collections4. Stag beetle larvae develop in and feed on decaying wood, playing a crucial role in forest ecosystems by promoting wood decomposition, nutrient recycling, and vegetation regeneration5,6. Adults of many species are nocturnal and primarily feed on tree sap and fermenting fruits4,7. Due to their ecological role and sensitivity to habitat changes, lucanid beetles are considered reliable bioindicators of forest matter cycling and ecosystem health8.
These beetles are distributed globally, occurring on all continents except Antarctica and inhabiting a diverse array of ecosystems, including forests, grasslands, and deserts9. The Lucanidae family is considered one of the most basal lineages within the superfamily Scarabaeoidea, underscoring its significant evolutionary importance10,11. Current research on stag beetles has primarily focused on taxonomy and phylogenetic relationships, drawing on nuclear gene fragments and mitochondrial multi-gene sequences12. High-quality genomic data are essential for gaining deeper insights into the evolutionary placement of Lucanidae within Scarabaeoidea. As of April 2025, only six Lucanidae genomes have been deposited in the NCBI database. In contrast to the rapidly growing number of genome assemblies for other beetle families, the availability of high-quality genomes for Lucanidae remains limited, highlighting the urgent need for additional genome sequencing and assembly efforts in this group.
To deepen our understanding of Lucanidae evolution and ecological adaptations, we assembled a chromosome-level genome of Odontolabis cuvera (Boisduval, 1835) by integrating PacBio HiFi long reads, Illumina short reads, and Hi-C data. Comprehensive genome annotation was performed, including identifying repetitive elements, non-coding RNAs, and protein-coding genes. This high-quality reference genome marks a significant advancement in Lucanidae research and provides a valuable genomic resource for exploring this beetle family’s evolutionary history and ecological adaptations.
Methods
Sample collection and sequencing
A single female specimen of O. cuvera was collected in Yunnan Province, China, on 24 October 2024 for concurrent DNA and RNA sequencing. Muscle tissue was carefully extracted from the pronotum and posterior abdominal segments. The tissue was washed in phosphate-buffered saline for five minutes to eliminate external contaminants. It was then flash-frozen in liquid nitrogen for 20 minutes and subsequently stored at −80 °C until sequencing procedures were initiated.
Genomic DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen), and total RNA was isolated with TRIzol Reagent (Thermo Fisher Scientific), following the manufacturers’ standard protocols. Illumina TruSeq DNA PCR-Free Kit was used to construct PCR-free libraries, yielding 150 bp paired-end reads. Hi-C libraries were generated by formaldehyde cross-linking, followed by MboI digestion, end-repair, and purification steps, following a standard protocol13. Short-read data were generated using the Illumina NovaSeq. 6000 platform. A 20 kb SMRTbell library was constructed (PacBio SMRTbell Express Template Prep Kit 2.0) and sequenced in HiFi mode on a PacBio Sequel II system. Berry Genomics (Beijing, China) conducted all library preparations and sequencing. In total, our sequencing efforts generated 160.95 Gb of data, including 36.70 Gb of PacBio HiFi long reads (61.02× coverage), 56.09 Gb of Illumina short reads (93.26×), and 58.56 Gb of Hi-C data (97.36×) (Table 1). PacBio HiFi sequencing generated reads with a scaffold N50 of 15.88 kb and an average read length of 15.93 kb.
Genome assembly
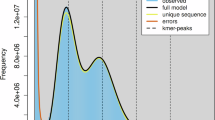
Raw Illumina reads were processed for quality control using BBTools v38.8214. Duplicate reads were first removed with “clumpify.sh”. Subsequently, bbduk.sh was applied to trim low-quality bases and adapter sequences according to strict quality criteria. This process involved discarding reads with Q < 20, removing reads with >5 Ns, trimming poly-A/G/C tails longer than 10 bp, and correcting overlapping paired reads. We conducted a k-mer-based genome survey analysis using GenomeScope v2.015 to estimate the genome size, heterozygosity, and repetitive sequence content of the O. cuvera genome. The estimated genome size ranged from 900.52 to 906.45 Mb, with repetitive elements comprising approximately 37.18–37.19% of the total genome. The analysis also revealed a heterozygosity rate of 1.13–1.39%, indicating a moderately high level of genetic diversity (Fig. 1).
The primary genome assembly of O. cuvera was performed using PacBio HiFi long reads with Hifiasm v0.19.816, applying default parameters. To eliminate redundant heterozygous sequences, Purge_Dups v1.2.517 was employed with a haploid cutoff value of 70 to identify and remove haplotigs effectively. Following quality control, Hi-C reads were aligned to the draft assembly using Juicer v1.6.218. Chromosome-level scaffolding was carried out with 3D-DNA v18092219, anchoring the primary contigs into chromosome-scale assemblies. The resulting genome assembly was meticulously reviewed, and any potential misassemblies were manually corrected using Juicebox v1.11.0818. To detect potential contaminants, we employed MMseqs. 2 v11.120 to conduct BLASTN-like searches against both the NCBI nucleotide and UniVec databases. Additional screening for vector contamination was performed using blastn (BLAST + v2.11.0)21 against the UniVec database. Sequences with over 90% identity to entries in either database were flagged as potential contaminants, while those with 80–90% identity underwent further verification through online BLASTN searches against the NCBI nucleotide database. Suspected bacterial and fungal contaminants were subsequently removed from the assembled sequences. The final O. cuvera genome assembly achieved chromosome-level resolution, with a total size of 908.07 Mb, comprising 66 scaffolds and 147 contigs, and a GC content of 32.65% (Table 2). A total of 81 gaps were present in the assembly. The scaffold and contig N50 values were 65.36 Mb and 16.39 Mb, respectively. In total, 99.58% of the assembled sequence (904.22 Mb) was successfully anchored to 14 chromosomes, which were ordered by descending length and ranged from 49.09 Mb to 94.68 Mb (Table 3; Figs. 2, 3).
Genome characteristics of Odontolabis cuvera. The circular genome plot displays, from the outermost to the innermost ring: (1) chromosome length, (2) GC content, (3) gene density, and (4) the distribution of major transposable elements, including DNA transposons, SINEs, LINEs, LTR retrotransposons, and simple repeats.
Genome annotation
To characterize repetitive elements in the O. cuvera genome, we performed de novo repeat annotation using RepeatModeler v2.0.422, incorporating the “-LTRStruct” pipeline to enhance the identification of LTR retrotransposons. The resulting repeat library was merged with RepBase-2023090923 and Dfam v3.524 to construct a comprehensive custom repeat database. RepeatMasker v4.1.225 was then employed to identify and mask repetitive sequences by aligning the genome against this integrated library. The RepeatMasker analysis revealed that approximately 481.28 Mb, accounting for 53.00% of the genome, consists of repetitive sequences. These include 233.31 Mb (25.69%) of unclassified repeats, 119.93 Mb (13.19%) of DNA transposons, 86.65 Mb (9.55%) of LINEs, 32.94 Mb (3.63%) of LTRs, and 5.86 Mb (0.65%) of simple repeats, along with additional repeat categories (Table 4).
Non-coding RNAs (ncRNAs) in the O. cuvera genome were annotated using Infernal v1.1.226 against the Rfam v14.1027 database, while tRNAscan-SE v2.0.928 was employed to predict transfer RNAs (tRNAs). In total, 1,219 ncRNAs were identified, including 4 long non-coding RNAs (lncRNAs), 64 ribozymes, 93 small nuclear RNAs (snRNAs), 99 microRNAs (miRNAs), 507 tRNAs, and 222 ribosomal RNAs (rRNAs) (Table 4).
The annotation of protein-coding genes in O. cuvera was conducted using MAKER v3.01.0329, an annotation pipeline that integrates multiple sources of evidence to produce high-confidence gene models. Three primary lines of evidence were incorporated: (1) transcriptomic evidence derived from RNA-seq reads aligned with HISAT2 v2.2.130 and assembled using StringTie v2.1.631; (2) ab initio predictions from BRAKER v2.1.632, incorporating both GeneMark-ES/ET/EP v4.68_lic33 and AUGUSTUS v3.4.034 pipelines trained on RNA-seq alignments and OrthoDB v1135 reference proteins; and (3) homology-based predictions generated by GeMoMa v1.936, leveraging protein sequences from five reference species: Drosophila melanogaster37 (GCF_000001215.4), Apis mellifera38 (GCA_003254395.2), Coccinella septempunctata39 (GCA_907165205.1), Prosopocoilus inquinatus40 (GCA_036172665.1), and Tribolium castaneum41 (GCA_031307605.1) (Table 5). The outputs from BRAKER and GeMoMa were merged and provided as ab initio input to the MAKER pipeline. A total of 21,798 predicted protein sequences were identified, reflecting that many genes produce multiple transcript variants. When considering only the longest transcript for each gene, the O. cuvera genome contained 18,332 predicted protein-coding genes, with an average gene length of 10,552.3 bp. Genes exhibited a mean structure of 5.4 exons, 4.4 introns, and 5.2 coding sequences (CDSs). Average exon length was 314.6 bp, while introns and CDSs measured 2,101.4 bp and 262.9 bp, respectively (Table 4). Gene set completeness was evaluated using BUSCO with the insecta_odb10 dataset (n = 1,367). The annotated protein-coding gene set exhibited 98.8% completeness, including 1,350 (97.7%) single-copy orthologs, 15 (1.1%) duplicated genes, 4 (0.3%) fragmented genes, and 13 (0.9%) missing genes. These results demonstrate that the gene annotations for O. cuvera are both comprehensive and of high quality.
Gene functional annotation was conducted using DIAMOND v2.0.11.142 in sensitive mode (–more-sensitive -e 1e-5) to align predicted protein sequences against the UniProtKB database. To further assign Gene Ontology (GO) terms, identify metabolic pathways (KEGG and Reactome), and annotate protein domains, we employed eggNOG-mapper v2.0.143 and InterProScan v5.53-87.044. The InterProScan analysis incorporated five databases: Pfam45, SMART46, SUPERFAMILY47, Gene3D48, and CDD49. Outputs from all tools were integrated to generate comprehensive functional annotations. In total, 16,972 genes were annotated with UniProt entries, 11,405 were assigned GO terms, 5,467 were mapped to KEGG pathways, 3,096 were associated with Enzyme Commission numbers, and 15,008 were classified into Clusters of Orthologous Groups (COG). Additionally, genome-wide distributions of repeat elements, gene density, and GC content across individual pseudochromosomes were visualized using TBtools50.
Data Records
The raw sequencing data and genome assembly of Odontolabis cuvera are publicly available through the National Center for Biotechnology Information (NCBI). The sequencing datasets, including Hi-C (SRR3279340551), transcriptome (SRR3183488052), Illumina short reads (SRR3183488153), and PacBio HiFi long reads (SRR3183488254), are publicly available under their respective accession numbers. The final genome assembly is available under NCBI accession GCA_049462965.155. Genome annotation files, including repeat element profiles, gene structure predictions, and functional annotations, are available via Figshare56.
Technical Validation
To evaluate the quality of the Odontolabis cuvera genome assembly, two complementary approaches were employed. First, genome assembly completeness was assessed using BUSCO v5.0.457 with the Insecta gene set (n = 1,367), revealing a high completeness score of 99.1%, with 98.3% single-copy, 0.8% duplicated, 0.3% fragmented, and 0.6% missing BUSCOs. Second, assembly accuracy was verified by mapping PacBio, Illumina, and RNA-seq reads to the final assembly using Minimap2 v2.2358 and SAMtools v1.959, achieving mapping rates of 99.99%, 88.28%, and 97.86%, respectively. These results demonstrate the high completeness and accuracy of the O. cuvera genome assembly.
Code availability
No specific script was used in this work. All commands and pipelines used in data processing were executed according to the manual and protocols of the corresponding bioinformatic software.
References
Fujita, H. The Lucanid Beetles of the World. Mushi-sha, Tokyo. (2010).
Inoue, A. & Hasegawa, E. Effect of morph types, body size and prior residence on food-site holding by males of the male-dimorphic stag beetle Prosopocoilus inclinatus (Coleoptera: Lucanidae). J Ethol. 31, 55–60 (2013).
Gotoh, H. et al. Developmental link between sex and nutrition; doublesex regulates sex-specific mandible growth via juvenile hormone signaling in stag beetles. PLoS Genet. 10, e1004098 (2014).
Kim, S. I. & Farrell, B. D. Phylogeny of world stag beetles (Coleoptera: Lucanidae) reveals a Gondwanan origin of Darwin’s stag beetle. Mol Phylogenet Evol. 86, 35–48 (2015).
Songvorawit, N., Butcher, B. A. & Chaisuekul, C. Decaying Wood Preference of Stag Beetles (Coleoptera: Lucanidae) in a Tropical Dry-Evergreen Forest. Environ. Entomol. 6, 1322–1328 (2017).
Chen, D., Cao, L. J., Zhao, J. L., Wan, X. & Wei, S. J. Geographic patterns of Lucanus (Coleoptera: Lucanidae) species diversity and environmental determinants in China. Ecol Evol. 10, 13190–13197 (2020).
Tanahashi, M., Matsushita, N. & Togashi, K. Are stag beetles fungivorous? J Insect Physiol. 55, 983–988 (2009).
Tanahashi, M., Ikeda, H. & Kubota, K. Elementary budget of stag beetle larvae associated with selective utilization of nitrogen in decaying wood. Sci Nat. 105, 33 (2018).
Kim, E. et al. Taxonomic note of the family Lucanidae (Coleoptera: Scarabaeoidea) in Cambodia. J Asia-Pac Entomol. 28, 102383 (2025).
Beaven, R., Denholm, B., Fremlin, M. & Scaccini, D. Evidence for the independent evolution of a rectal complex within the beetle superfamily Scarabaeoidea. Arthropod Struct Dev. 84, 101406 (2025).
McKenna, D. D. et al. The evolution and genomic basis of beetle diversity. Proc. Natl. Acad. Sci. USA. 116, 24729–24737 (2019).
Zeng, L. et al. Comparative mitochondrial genomics of five Dermestid beetles (Coleoptera: Dermestidae) and its implications for phylogeny. Genomics. 113, 927–934 (2021).
Belton, J. M. et al. Hi-C: A comprehensive technique to capture the conformation of genomes. Methods. 58, 268–276 (2012).
Bushnell, B. BBtools. Available online: https://sourceforge.net/projects/bbmap/ (accessed on 1 October 2022) (2014).
Ranallo-Benavidez, T. R., Jaron, K. S. & Schatz, M. C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 11, 1432 (2020).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 18, 170–175 (2021).
Guan, D. et al. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 36, 2896–2898 (2020).
Durand, N. C. et al. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 3, 95–98 (2016).
Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 356, 92–95 (2017).
Steinegger, M. & Soding, J. MMseqs. 2 enables sensitive protein sequence searching for the analysisof massive datasets. Nat. Biotechnol. 35, 1026–1028 (2017).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Flynn, J. M. et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA. 117, 9451–9457 (2020).
Bao, W., Kojima, K. K. & Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. Dna. 6, 11 (2015).
Hubley, R. et al. The Dfam database of repetitive DNA families. Nucleic Acids Res. 44, D81–D89 (2016).
Smit, A. F. A., Hubley, R. & Green, P. RepeatMasker Open-4.0. Available online: http://www.repeatmasker.org (accessed on 1 October 2022) (2013–2015).
Nawrocki, E. P. & Eddy, S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 29, 2933–2935 (2013).
Griffiths-Jones, S. et al. Rfam: annotating noncoding RNAs in complete genomes. Nucleic Acids Res. 33, D121–124 (2005).
Chan, P. P. & Lowe, T. M. TRNAscan-SE: Searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962, 1–14 (2019).
Holt, C. & Yandell, M. MAKER2: An annotation pipeline and genome-database management tool for second-generation genome projects. Bmc Bioinformatics. 12, 491 (2011).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 12, 357–360 (2015).
Kovaka, S. et al. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20, 278 (2019).
Bruna, T., Hoff, K. J., Lomsadze, A., Stanke, M. & Borodovsky, M. BRAKER2: Automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. Nar Genom. Bioinform. 3, lqaa108 (2021).
Bruna, T., Lomsadze, A. & Borodovsky, M. GeneMark-EP: Eukaryotic gene prediction with self-training in the space of genes and proteins. Nar Genom. Bioinform. 2, lqaa26 (2020).
Stanke, M., Steinkamp, R., Waack, S. & Morgenstern, B. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 32, W309–W312 (2004).
Kriventseva, E. V. et al. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 47, D807–D811 (2019).
Keilwagen, J., Hartung, F., Paulini, M., Twardziok, S. O. & Grau, J. Combining RNA-seq data and homology-based gene prediction for plants, animals and fungi. Bmc Bioinformatics. 19, 189 (2018).
Hoskins, R. A. et al. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome research. 25, 445–458 (2015).
Gibbs, R. A. et al. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 443, 931–949 (2006).
Crowley, L. The genome sequence of the seven-spotted ladybird, Coccinella septempunctata Linnaeus, 1758. Wellcome open research. 6, 319 (2021).
Pang, B., Zhan, Z. & Wang, Y. A chromosome-level genome assembly of Prosopocoilus inquinatus Westwood, 1848 (Coleoptera: Lucanidae). Sci Data. 11, 808 (2024).
Herndon, N. et al. Enhanced genome assembly and a new official gene set for Tribolium castaneum. BMC Genomics. 21, 47 (2020).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 12, 59–60 (2015).
Huerta-Cepas, J. et al. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol. Biol. Evol. 34, 2115–2122 (2017).
Finn, R. D. et al. InterPro in 2017—Beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199 (2017).
El-Gebali, S. et al. The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 (2019).
Letunic, I. & Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496 (2018).
Wilson, D. et al. SUPERFAMILY—Sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 37, D380–D386 (2009).
Lewis, T. E. et al. Gene3D: Extensive Prediction of Globular Domains in Proteins. Nucleic Acids Res. 46, D1282 (2018).
Marchler-Bauer, A. et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203 (2017).
Chen, C. et al. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 13, 1194–1202 (2020).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR32793405 (2025).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR31834880 (2025).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR31834881 (2025).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR31834882 (2025).
NCBI Assembly https://identifiers.org/ncbi/insdc.gca:GCA_049462965.1 (2024).
Zhu, M. Genome annotation (repeats and protein-coding genes). figshare. Dataset. https://doi.org/10.6084/m9.figshare.28787375.v1 (2025).
Waterhouse, R. M. et al. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 35, 543–548 (2018).
Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics. 34 (2018).
Dudchenko, O. et al. Twelve years of SAMtools and BCFtools. GigaScience. 10(2), giab008 (2021).
Acknowledgements
This study was supported by grants from Henan Science and Technology Research Project (252102320222).
Author information
Authors and Affiliations
Contributions
Y.J. and H.Y. contributed to the research design. Z.M., Z.J. and H.Y. collected the samples. Z.M. analyzed the data. Z.M., and H.Y. wrote the draft manuscript and revised the manuscript. All co-authors contributed to this manuscript and approved it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhu, M., Han, Y., Zhang, J. et al. A chromosomal-level genome assembly of Odontolabis cuvera Hope, 1842 (Coleoptera: Lucanidae). Sci Data 12, 1258 (2025). https://doi.org/10.1038/s41597-025-05613-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05613-5