Abstract
Qihe gibel carp (Carassius gibelio var. Qihe) is a local population of natural gynogenetic amphitriploid (AAABBB) Carassius gibelio, and has high nutritional and economic value. In this study, we assemble a high-quality chromosome-level genome of Qihe gibel carp through DNBSEQ, PacBio HiFi, and Hi-C sequencing data. The resulting assembly consisted of 350 contigs with the full length of 1.607 Gb and 96.21% (1.515 Gb) of the assembled genome was successfully anchored to 50 chromosomes, with a contig N50 of 28.97 Mb and a scaffold N50 of 29.84 Mb. Repeated sequences accounting for 43.72% (732.494 Mb) of the total were also identified, and gene prediction revealed 46,131 protein-coding genes with an annotation ratio of 96.48%. Furthermore, Benchmarking Universal Single-Copy Orthologue (BUSCO) analysis demonstrated that the genome assembly achieved high completeness, with a score of 97.66%. This high-quality chromosome-level genome lays the foundation for molecular biology research as well as molecular breeding and evolutionary studies of Qihe gibel carp in the future.
Similar content being viewed by others
Background & Summary
The genus Carassius play crucial roles in aquaculture. They are rare vertebrates which exhibit various ploidy levels and reproductive modes, including the sexual amphidiploid (allotetraploid, AABB) crucian carp Carassius auratus (C. auratus) and the unisexual amphitriploid (AAABBB) gibel carp Carassius gibelio (C. gibelio)1,2,3,4. Qihe gibel carp (C. gibelio var. Qihe) is a geographic population of C. gibelio that is cultured in the middle and lower reaches of the Qihe River in Henan Province, China; it is also well-known as the “double-backed crucian carp” due to its wide and thick back5,6. Besides, Qihe gibel carp is a rare unisexual amphitriploid (AAABBB) freshwater fish with gynogenesis ability7. Because of numerous obstacles present in the pairing and equal separation of three sets of chromosomes during meiosis and gametogenesis, amphitriploid organism is very rare in nature, especially in vertebrates8. However, Qihe gibel carp can overcome reproductive barriers through gynogenesis, which makes them valuable for studies of species evolution and fish genetic breeding. Moreover, Qihe gibel carp also has high nutritional and economic value, and belongs to precious protected germplasm resource in China9. However, the lack of high-quality reference genomes of Qihe crucian carp has hindered deeper gene function and high-quality breeding of new varities derived from this fish germplasm resources.
In recent years, genome sequencing technology has developed rapidly. The long-read sequencing technologies (PacBio HiFi or Oxford Nanopore Technologies, ONT) have been widely applied in chromosome-level genomes assembly of plants and animals, such as fish species. Compared to the second-generation sequencing technologies, long-read sequencing technology allows the completion of the assembly of the chromosome-level genome of fish and also contributes to the continuous development of structural and functional genomics research processes10. Therefore, studies of aquaculture species have progressed from gene-level research to whole genome-level research.
Using genome sequencing technology, researchers have conducted genetic evolution analysis of different fish species and revealed the evolutionary mechanisms of polyploid fish. For example, Li et al. (2021) conducted genome assembly and genome annotation of all-tetraploid common carp and goldfish, and sequenced three common carp strains11. Their results identified candidate genes related to growth and survival rate, revealed the evolutionary mechanism of the parallel subgenome structure and divergent expression evolution of common carp and goldfish, and elucidated the geographic genome structure and domestication of common carp. Wang et al. (2022) assembled the genome of one amphitriploid (AAABBB) gibel carp (C. gibelio) and one amphidiploid crucian carp/goldfish (C. auratus))12. Combined with comparative genomic analysis and cytological observations, they reported some evidence of genomic variation to promote the gynogenesis of C. gibelio, and they provided new insights into the evolutionary mechanism of successful reproduction in unisexual polyploid vertebrates. In another study, Xu et al. (2023) revealed the origin and subsequent sub-genome evolution patterns following three independent allopolyploidy events by assembling the high-quality genomes of 21 cyprinids13.
Qihe gibel carp has important breeding value as well as research value for understanding genome evolution. However, the genome of Qihe gibel carp has not been reported to date. Although Wang et al. (2022) assembled the genome of gibel carp12, in view of the fact that Qihe crucian carp as an important geographical population and its genetic isolation, and some scientists believe that the doubling event of crucian carp is independent, it is necessary to de nove sequence and assemble the chromosome level genome of Qihe crucian carp. Therefore, we used PacBio HiFi and High-throughput Chromosome Conformation Capture (Hi-C) technologies to sequence and assemble a high-quality chromosome-level genome of Qihe gibel carp. The resulting assembly consisted of 350 contigs with the full length of 1.607 Gb and 96.21% (1.515 Gb) of the assembled genome was successfully anchored to 50 chromosomes, with a contig N50 of 28.97 Mb and a scaffold N50 of 29.84 Mb. Benchmarking Universal Single-Copy Orthologue (BUSCO) analysis demonstrated that the genome assembly achieved high completeness, with a score of 97.66%. Genomic annotation was performed through de novo gene prediction, homology, and transcriptome-based prediction. Repeated sequences accounting for 43.72% (732.494 Mb) of the total were also identified, and gene prediction revealed 46,131 protein-coding genes with an annotation ratio of 96.48%. This study lays the foundation for the construction of genetic maps, the evaluation of germplasm resources, molecular breeding, and genetic classification studies of Qihe gibel carp in the future.
Methods
Sample collection and sequencing
The Qihe gibel carp used in this experiment were collected from Qihe gibel carp breeding base of Anyang Institute of Technology (Anyang, Henan, China). All experiments complied with institutional animal care guidelines and were approved by the Animal Care Committee of Anyang Institute of Technology. Muscle tissue from female Qihe gibel carp was used to extract DNA following the manufacturer’s instructions of the FastPure DNA Isolation Mini Kit (Vazyme, Nanjing, China). The purity and concentration of the extracted DNA were assessed using a NanoDrop 2000 system (Thermo Scientific, Waltham, MA, USA) and gel electrophoresis.
The short-read library was constructed and sequenced using a DNBSEQ-T7 platform in BGI Co., Ltd. A total of 196.64 Gb of clean data was obtained. We employed the SMRT Bell Express Template Prep Kit 2.0 (Pacific Biosciences, Menlo Park, CA, USA) to construct a PacBio SMRT Bell library, which was used for SMRT sequencing on the PacBio platform in BGI Co., Ltd. Using CCS workfow13 version 4.0.0, the raw data were converted into high-quality HiFi reads. In total, 228.01 Gb of HiFi reads were generated.
The Hi-C library was constructed using the same tissues, and it was sequenced on the DNBSEQ-T7 platform. A total of 406.46 Gb clean data was generated. For ISO-seq, muscle tissue of female Qihe gibel carp was also used to extract RNA following the manufacturer’s instructions of the modified CTAB extraction method. The PacBio ISO-seq library was constructed and sequenced using the PacBio Sequel II platform in BGI Co., Ltd. In total 49.78 Gb of clean data were obtained (Table 1).
Genome survey and assembly
To survey the genome, we assessed several important characteristics, including repeated sequence, genome size, and heterozygosity. Based on MGI-DNBSEQ (https://en.mgi-tech.com/products/), the k-mer (k = 21) frequencies were analyzed using GCE version 1.0.014. The obtained k-mer frequency information was used to evaluate genome heterozygosity, repeatability, and genome size using GenomeScope 2.0. The calculated haploid genome scale of Qihe gibel carp was 1.52 Gb, with a repeated content of 40.08% and a heterozygosity of 2.20% (Fig. 1). Next, circular consensus sequencing reads were assembled using Hifiasm version 0.19.6 (https://github.com/chhylp123/hifiasm) with default parameters. We obtained 350 contigs with a total length of 1.607 Gb and a contig N50 of 28.97 Mb, which constituted the genome assembly. The Hi-C clean reads were mapped into reference genome using BWA15 (version 0.7.12-r1039) with default parameters, which generated the uniquely aligned read pairs needed to evaluate and obviate the invalid read pairs. A length of 2,778,181,521 base pair sequences were used for scaffold correction to further cluster, order, and orient contigs through Juicer16 (version 1.6) with default parameters, and then the assembled contigs were anchored to chromosomes using 3D-DNA software17 (version 180,922) with -r 0.parameters (https://github.com/theaidenlab/3d-dna) Based on JuiceBox software18 (version 1.11.08) with default parameters, the interaction graph was visualized to perform manual correction. Ultimately, there were 316 scaffolds of 1.515 Gb in length with a scaffold N50 of 29.84 Mb anchored to 50 chromosomes, which represented 96.21% of the contigs in length.
Repeat sequence prediction
Repeat sequences play a key role in the gene regulatory network. They include interspersed repeats and tandem repeats, which may mainly be transposable elements (TEs), and they play an indispensable role in the evolution, inheritance, and variation of life and in gene expression and transcriptional regulation. At present, the methods for annotating repeat sequences are mainly de novo prediction and homologous comparison, which are based on the RepBase library (http://www.girinst.org/repbase)19. RepeatMask (version open-4.0.9) with default parameters20 and RepeatProteinMask (version open-4.0.9) with -nolow-no_is-norna parameters20 software are used to identify sequences that are similar to known repeat sequences. In our study, de novo prediction was mainly achieved through RepeatModeler (version open-1.0.11) with default parameters21 and LTR_Finder (version 1.0.7) with default parameters22 software, which first constructed a de novo repeat sequence library that RepeatMask software used to predict the repeat sequences. Tandem Repeats Finder (version 4.09) with default parameters23 was also used to find tandem repeat sequences. In total 731.56 Mb of repeat sequences were annotated, which accounted for 46.46% of the genome assembly, similar with 728.98 Mb (45.85) of repeat sequences in C. gibelio12. Additionally, 691.57 Mb of TEs were identified, accounting for 43.92% of the genome assembly (Tables 2, 3, Fig. 2).
Protein-coding gene prediction and functional annotation
Gene prediction of the Qihe gibel carp genome was performed using a combination of de novo-based, transcriptome-based, and homolog-based gene prediction methods. The de novo-based prediction was performed using AUGUSTUS24 (version 3.3.2) and Genscan25 software. Protein sequences from Carassius auratus (PRJNA546444), Carassius gibelio (PRJNA546443), Danio rerio (GCF_000002035.6), and Poropuntius huangchuchieni26 were compared with the Qihe gibel carp genome to establish homology-based prediction using Exonerate27 version 2.2.0 and Liftoff28 (version 1.6.3) with default parameters.
Based on the third-generation sequencing ISO-seq data, the Iso-seq reads were aligned to the genome sequence using HISAT2 version 2.1.0 with default parameters29. The transcripts were predicted using Stringtie (version 1.3.5) with default parameters30, and the intron sequences of all samples were extracted for subsequent auxiliary annotation by TransDecoder (version 5.5.0) (https://github.com/TransDecoder/TransDecoder). In addition, the integrated gene database was successfully constructed using MAKER2 (version 2.31.10) with default parameters31 and HiFAP with default parameters. During the database processing, variable splicing was removed and the transcriptome-specific genes were added. Totally, the assembled genome was improved by filtering out the parts with only a single coding sequence and corresponding genes without functional annotations.
In total, 46,131 protein-coding genes were identified from the Qihe gibel carp genome (Table 4). Subsequently, the protein-coding genes were functionally annotated in Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), Non-redundant (NR), EuKaryotic Orthologous Groups (KOG), InterPro, Swiss-Prot, and TrEMBL databases using Diamond (version 2.0.14) with--evalue 1e-05 parameters32. A total of 44,509 protein-coding genes were annotated, accounting for 96.48% of the total identified protein-coding genes (46,131) (Table 4).
There were 49,299 non-coding RNAs in the Qihe gibel carp genome (Table 5). Non-coding RNAs mainly include transfer RNA, ribosomal RNA, micro RNA, and small nuclear RNA. Transfer RNAs were identified based on their structural characteristics using tRNAscan-SE (version 1.3.1) with default parameters33. Ribosomal RNAs were identified using BLASTN software. Micro RNAs and small nuclear RNAs were annotated using Rfam (version 14.8) with cmscan–rfam–nohmmonly parameters34.
Genomic synteny analysis
The genomic synteny analysis between Qihe gibel carp and C. auratus or C. gibelio was performed using WGDI (version 0.5.6) with score = 100, evalue = 1.0E-5, ks_col = ks_NG86 parameters35. First, all versus and alignment of the protein sequences from both species was performed using BLASTP version 2.11.0 + (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Second, gene position and chromosome length information were extracted. Finally, a graphical presentation of genome synteny analysis was produced using JCVI36 (version 1.1.22). The results showed that there was no fission event between Qihe gibel carp and C. auratus or C. gibelio, which implies high concordance of the genomes between Qihe gibel carp and both of the other fishes and also indirectly confirms the credibility of the genome assembly of Qihe gibel carp (Fig. 3).
Data Records
The ISO-seq sequencing data by PacBio Sequel II, genomic survey sequencing data by DNBSEQ-T7, genomic PacBio HiFi sequencing data by PacBio, and Hi-C sequencing data by DNBSEQ-T7 from Qihe gibel carp were deposited in the NCBI SRA database (accession number: SRP533627)37. The assembled Chromosome-level genome of Qihe gibel carp was submitted to NCBI (accession number: GCA_043790415.1)38. The annotated files for the Qihe gibel carp genome were deposited at figshare39.
Technical Validation
Benchmarking Universal Single-Copy Orthologue (BUSCO) is tool that is used to search for single-copy orthologs between species to evaluate the integrity and redundancy of one new assembled genome. In this study, the assembled results of the Qihe gibel carp genome were evaluated using BUSCO40 (version 5.3.1) with -m genome–limit 10 parameters. The complete BUSCO at contig-level assembly was 97.4%, while the complete BUSCO at chromosome-level assembly was 97.7%, similar with 98.16% of C. gibelio12, and these values indicate that a high-quality Qihe gibel carp genome was obtained in this study. In addition, 97.3% of all protein-coding genes and 97.1% of functionally annotated protein-coding genes were identified as complete (Table 6).
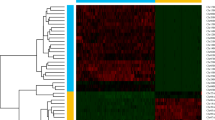
The genome assembled at the chromosome level was divided into bins in equal lengths of 100 kb using Hi-C data. The Hi-C read pair numbers covered between any two bins were counted as the signal intensity of the interaction between the two bins, and the data were used to create a heat map. The intensity of interactions located at diagonal positions was higher than that at non-diagonal positions in each group, suggesting that the interaction intensity was high between adjacent sequences at diagonal positions and weak between non-adjacent sequences at non-diagonal positions. These results were consistent with the principle of genome assembly assisted by Hi-C, illustrating that the genomic assembly was highly effective (Fig. 4).
Code availability
All data analyzing tools and software used in this study were performed following the instructions and guidelines. There was no custom code applied to analyze the data in our study.
References
Jiang, Y. G., Yu, H. X., Chen, B. D. & Liang, S. C. Biological effect of heterologous sperm on gynogenetic offspring in Carassius auratus gibelio. Acta Hydrobiol Sin. 8, 1–13 (1983).
Lu, M. et al. Changes in Ploidy Drive Reproduction Transition and Genomic Diversity in a Polyploid Fish Complex. Mol Biol Evol. 39, msac188 (2022).
Gui, J. F., Zhou, L. & Li, X. Y. Rethinking fish biology and biotechnologies in the challenge era for burgeoning genome resources and strengthening food security. Water Biology and Security. 1, 16 (2022).
Gui, J. F. Chinese wisdom and modern innovation of aquaculture. Water Biology and Security. 3, 100271 (2024).
Zhou, C., Li, B., Ma, L., Zhao, Y. & Kong, X. The complete mitogenome of natural triploid Carassius auratus in Qihe River. Mitochondrial DNA A DNA Mapp. Seq Anal. 27, 605–606 (2016).
Li, F. B. & Gui, J. F. Clonal diversity and genealogical relationships of gibel carp in four hatcheries. Anim Genet. 39, 28–33 (2008).
Jiang, H. et al. Response of Acid and Alkaline Phosphatase Activities to Copper Exposure and Recovery in Freshwater Fish Carassius auratus gibelio var. Life Science Journal. 9, 233–245 (2012).
Lu, M. et al. Regain of sex determination system and sexual reproduction ability in a synthetic octoploid male fish. Sci China Life Sci. 64, 77–87 (2021).
Gao, Y. et al. Blood biochemistry profile of Qihe gibel carp Carassius auratus in different aquaponic systems. Environ Sci Pollut Res Int. 27, 42898–42907 (2020).
Rhie, A. et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature. 592, 737–746 (2021).
Li, J. T. et al. Parallel subgenome structure and divergent expression evolution of allo-tetraploid common carp and goldfish. Nat Genet. 53, 1493–1503 (2021).
Wang, Y. et al. Comparative genome anatomy reveals evolutionary insights into a unique amphitriploid fish. Nat Ecol Evol. 6, 1354–1366 (2022).
Xu, M. R. et al. Maternal dominance contributes to subgenome differentiation in allopolyploid fishes. Nat Commun. 14, 8357 (2023).
Liu, B. H. et al. Estimation of genomic characteristics by analyzing K-mer frequency in de novo genome projects. Quant. Biol. 35, 62–67 (2013).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25, 1754–1760 (2009).
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments[J]. Cell systems. 3, 95–98 (2016).
Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds[J]. Science. 356, 92–95 (2017).
Durand, N. C. et al. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Syst. 3, 99–101 (2016).
Jurka, J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110, 462–467 (2005).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 4, 3 (2009).
Flynn, J. M. et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci USA. 117, 9451–9457 (2020).
Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, 265–268 (2007).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, 435–439 (2006).
Burge, C. & Karlin, S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1, 78–94 (1997).
Chen, L. et al. Chromosome-level genome of Poropuntius huangchuchieni provides a diploid progenitor-like reference genome for the allotetraploid Cyprinus carpio. Mol Ecol Resour. 21, 1658–1669 (2021).
Slater, G. S. & Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 6, 1–11 (2005).
Alaina, S. & Salzberg, S. L. Liftoff: accurate mapping of gene annotations. Bioinformatics. 37, 1639–1643 (2021).
Kim, D. et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat biotechnol. 37, 907–915 (2019).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat biotechnol. 33, 290–295 (2015).
Holt, C. & Yandell, M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 12, 1–14 (2011).
Buchfink, B., Reuter, K. & Drost, H. G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nature methods. 18, 366–368 (2021).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Griffiths-Jones, S. et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Research. 33, 121–124 (2005).
Sun, P. et al. WGDI: A user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. Mol Plant. 15, 1841–1851 (2022).
Tang, H. et al. Synteny and Collinearity in Plant Genomes. Springer Netherlands (2008).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP533627 (2024).
NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_043790415.1 (2024).
Lian, K. Q. The annotation data of Qihe gibel carp genome. figshare https://doi.org/10.6084/m9.figshare.27299880 (2024).
Simão, F. A. et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 19, 3210–3212 (2015).
Acknowledgements
This work is supported by National Key R&D Program of China (2023YFD2400300), Science and Technology Research Project of Henan Province (242102110065), Special Fund for Henan Agriculture Research System (HARS-22-16-G4), Science and Technology Research Project of Anyang City (2023C01NY013), and Central Public-interest Scientific Institution Basal Research Fund, CAFS (2024JC0105). We appreciate the assistance from Xuejun Li and Jianxin Feng during the sampling. We thank Wuhan Onemore-tech Co., Ltd. for their assistance with genome sequencing and analysis.
Author information
Authors and Affiliations
Contributions
R.P., J.G. and Y.K. conceived the project; K.L., J.S., J.M., X.L. and Y.L. performed the experiments; K.L. and M.D. did the bioinformatic analyses; R.P., K.L., M.D., M.L. L.Z., J.W. and J.S. evaluated the data; K.L. and J.S. wrote the manuscript. The final submission received recognition and approval from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lian, K., Shan, J., Ma, J. et al. Chromosome-level genome assembly of Qihe gibel carp. Sci Data 12, 1290 (2025). https://doi.org/10.1038/s41597-025-05636-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05636-y