Abstract
The root-zone microbiome of wheat, encompassing root, rhizosphere and bulk soil communities, harbors diverse diazotrophs that critically enhance wheat nitrogen use efficiency, promote wheat growth, and improve abiotic stress tolerance. Despite their agricultural significance, the composition and temporal dynamics of these beneficial diazotroph communities in saline-alkali ecosystems remain poorly resolved, particularly under the stresses of aridity and soil salinity. Systematic profiling of wheat-associated diazotroph ecology is imperative for developing innovative solutions to enhance wheat resilience and advance sustainable agriculture. This study provides detailed microbiome data from wheat root-soil interface, consisting of root, rhizosphere and bulk soil samples collected from saline-alkali fields in the water-scarce Bohai Sea region of China. Through high-throughput sequencing of nifH gene, we generated an extensive dataset with an average of 60,265 high-quality reads per sample. The dataset provides a valuable resource for uncovering the recruitment strategies of wheat root-enriched diazotrophs to adapt to varying salinity stress and facilitating the development of novel microbiome based strategies for sustainable agricultural practices in saline-alkali ecosystems.
Similar content being viewed by others
Background & Summary
Nitrogen (N) plays a vital role in agricultural production and plant growth. Although plants require substantial nitrogen for optimal growth, they cannot directly utilize atmospheric N2. Instead, N2 must be biologically converted into ammonia (NH3) by diazotroph communities through the action of the nitrogenase enzyme1. These beneficial microbes enhance plant growth through multiple mechanisms, including phosphate solubilization, root morphology modulation, osmotic regulation, increased siderophore production, enhanced solubilization activity, and stomatal regulation. However, the saline-alkali soils severely reduce the diversity of diazotroph community in the root-zone and lead to a deficiency in soil nitrogen contents2. Soil salinization adversely affects plant growth and soil health, emerging as one of the major constraints on global agricultural productivity3. In the context of increasingly severe climate change and anthropogenic disturbances, the area of saline-alkali land continues to expand, and innovative approaches are urgently required to alleviate plant stress and boost the resilience of wheat crops in these challenging environments.
Diazotroph communities derived from diverse wheat habitats-including the root, rhizosphere and bulk soil, serve as main contributors to biological nitrogen fixation (BNF). This process is driven by highly diverse microorganisms and plays a pivotal role in enhancing wheat nitrogen use efficiency and exhibits remarkable capabilities in improving plant resilience to saline-alkali stress4. Emerging evidence indicates that rhizosphere microbial communities are particularly crucial for plant adaptation under saline-alkaline conditions5,6. These microbes enhance salt tolerance through multiple synergistic mechanisms: direct stress protection via biosynthesis of osmoprotectants (proline and poly-γ-glutamic acid) that simultaneously function as salinity stress signals7; metabolic compensation through photosynthetic enhancement and phytohormonal regulation (IAA-ethylene crosstalk) to offset energy demands during stress adaptation8; dual-function metabolites that simultaneously stimulate crop growth and elevate systemic resistance to saline-alkali stress through physiological modulation.
Accumulating evidence demonstrates that plant growth-promoting rhizobacteria inoculation can mitigate the detrimental effects of high salinity stress on wheat while enhancing growth performance9. Most diazotrophs have growth promotion potential in field environments. These beneficial relationships with wheat can reduce susceptibility to environmental salinity stresses, corresponding to traits that are likely to be essential for sustaining survival or facilitating adaptation in the rhizosphere microhabitats, thereby promoting wheat growth. These beneficial microbes not only improve soil nitrogen supply but also reduce soil pH and electrical conductivity, ultimately optimizing the rhizosphere microenvironment for wheat roots10. Haroon et al. demonstrated that diazotroph inoculation significantly enhances wheat growth in saline-alkaline environments while reducing dependence on nitrogen fertilizers11. When facing salinity stress, plants will recruit more diazotrophs to fix more nitrogen to improve plant tolerance to salinity stresses and bring about higher diazotroph community richness in root and rhizosphere soils. Single-strain inoculants, when introduced, often exhibit limited efficacy due to poor colonization efficiency and competition with indigenous soil microbial communities. This suggests that plants may derive greater benefits from functionally diverse microbial communities rather than individual strains12. Understanding the diversity of diazotroph community in the wheat rhizosphere could disentangle the complex interactions between the wheat grown in association with their root microbiota.
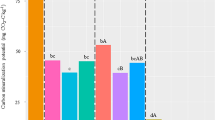
To evaluate the diversity of diazotroph community in wheat root zones of saline-alkali soils, by using high-throughput amplicon sequencing of the nifH gene fragments from 54 samples representing root, rhizosphere and bulk soil across different salinity levels (low: 221 μS/cm, medium: 383 μS/cm, high: 899 μS/cm, Table 1) and growth stages (April and May) in the water-scarce Bohai Sea region of China (Fig. 1). A total of 5,640,071 raw reads were obtained. After quality control, the median number of high-quality reads was 57,218, corresponding to 2,319 operational taxonomic units (OTUs) from the nifH region. The minimum sequencing depth is 5,781, the maximum sequencing depth is 104,584, the mean frequency is 60,265 (Fig. 2). The most abundant diazotrophic phylum Pseudomonadota (synonym Proteobacteria) remained consistent across all salinity levels, periods, and wheat root-soil interface, including root, rhizosphere and bulk soil, although relative abundance variations were observed (Fig. 3a), the top ten abundant diazotrophic families including Anaeromyxobacteraceae, Comamonadaceae, Desulfuromonadaceae, Azospirillaceae, Geobacteraceae, Nitrobacteraceae, Zoogloeaceae, Rhizobiaceae, Pseudomonadaceae, Paracoccaceae (Fig. 3b). Diazotroph community was clearly separated by microhabitats (root, rhizosphere and bulk soil) and sampling period (April and May) among all treatments shown visually as non-metric multidimensional scaling (NMDS) plot (Fig. 4). Multiple samples rarefaction curves were used to show that the sequencing depths of the samples are sufficient to represent the actual species richness (Fig. 5).
Comparison of diazotroph community structure in different samples. Non-metric multidimensional scaling (NMDS) based on Bray‒Curtis distance showing the profile of diazotroph community in wheat root (R), rhizosphere (RS), and bulk soil (BS). L mean low salinity, M mean medium salinity, H mean high salinity; 4 mean April, 5 mean May; BS mean bulk soil, RS mean rhizosphere soil, R mean root.
In summary, this study provides valuable data into the diazotroph community in wheat root zones (root, rhizosphere and bulk soil) under saline-alkaline conditions. This dataset offers microbial strategic guidance for saline soil reclamation and soil biodiversity conservation.
Methods
Site description and sample collection
The field experiment was located in a coastal saline zone in Huanghua County, Cangzhou city, China, on the western side of Bohai Bay (117°26′30″E, 38°28′35″N). Initially, root, rhizosphere and bulk soil samples were collected in April and May 2024. Within each field, wheat samples were carefully removed from the soil using a shovel. Root samples, together with adhering soil particles, were collected for each plant. Bulk soil samples were collected from the unplanted plots using a 4 cm diameter, 20 cm deep soil core sampler. Part of each soil sample was then sieved through a 2 mm mesh to remove visible organic matter and stones and dried the soil samples indoors to remove excess moisture for soil salt content measurement by conductivity method, while the remainder was stored at −20 °C for DNA extraction.
To process the root-associated samples, we used a protocol to obtain the microbiome living within the plant13. Briefly, 5 g of root tissue was transferred into a 50 mL conical flask containing 30 mL of sterile phosphate buffered saline (PBS). The mixture was shaken in a rocking incubator at 200 rpm for 20 min, the liquid was collected to obtain the rhizosphere sample, and the root samples were transferred to a new 50 mL conical flask. To isolate rhizosphere DNA, the liquid sample underwent centrifugation at 10,000 × g for 5 minutes. Subsequently, 500 mg of the dense pellet—comprising fine sediment and microorganisms—was transferred to a Lysing Matrix E tube (included in the DNeasy PowerSoil Pro Kit for soil sample processing). To obtain root microbiome from the root samples, the roots were rinsed three times with sterile ddH2O to completely remove surface soil, placed in a 50 ml Falcon tube containing 30 ml PBS and sonicated for 10 min using an ultrasonic cell disruptor at low frequency (30 s bursts followed by 30 s pauses). The sonicated roots were first rinsed three times with sterile water, followed by disinfection in 30 mL of 2% sodium hypochlorite for 5 minutes. After disinfection, the roots were again washed thoroughly with sterile water. To verify the removal of all surface microbes, PCR was conducted using the washed roots as a template. Prior to total genomic DNA extraction, liquid nitrogen was added to the root samples, which were then crushed to facilitate DNA isolation.
Soil physicochemical analysis
Soil properties were measured based on the methods described in a previous study14. The electrical conductivity (EC) of soil samples was measured in a 1:5 (wt/vol) mixture of moist soil and boiled deionized water using a DDS-307A conductivity meter (INASA Scientific Instrument Co., Ltd., China). Soil pH was assessed with an S220 Seven Compact pH meter (Mettler Toledo, Switzerland) at a soil: water ratio of 1:2.5 (dry weight/volume). Soil organic matter (SOM) content was determined via the dichromate oxidation method, and total nitrogen (TN) was measured using a modified Kjeldahl approach. TP was determined by alkali fusion combined with molybdenum-antimony-anti colorimetry. TK in soil was determined by alkali fusion combined with flame photometry or atomic absorption spectrometry (AAS). Alkali-hydrolyzable nitrogen (available potential nitrogen) in soil was determined using the diffusion method. Soil Olsen P and available K contents were extracted after 0.5 M NaHCO3 (pH = 8.5) and 1 M NH4OAc digestion and measured using the molybdenum blue method and flame spectrophotometry method (FP640, INASA, China), respectively.
Molecular analyses and bioinformatic analyses
Total genomic DNA was extracted using DNeasy PowerSoil Pro Kit according to the manufacturer’s protocol. The concentration and purity of the extracted DNA were checked using a NanoDrop One system and stored at −20 °C until molecular analysis. Primers nifHFor/nifHRev (5′- TGCGAYCCSAARGCBGACT -3′/5′- ATSGCCATCATYTCRCCGGA -3′) of the nifH gene were used for amplicon sequencing of the diazotroph community. PCR amplification was carried out using the Q5 High-Fidelity DNA Polymerase kit (NEB, #M0491L) in a 25-μL reaction volume. The thermal cycling conditions were as follows: initial denaturation at 98 °C for 2 minutes, followed by 30 cycles consisting of denaturation at 98 °C for 30 seconds, annealing at 59.5 °C for 30 seconds, and extension at 72 °C for 45 seconds, with a final extension step at 72 °C for 5 minutes. The resulting amplicons were then subjected to Illumina paired-end sequencing on the NovaSeq platform.
Microbiome bioinformatics were mainly performed with Vsearch and Quantitative Insights Into Microbial Ecology (QIIME 2)15. Briefly, raw sequence data were demultiplexed using the demux plugin followed by primers cutting with cutadapt plugin16. Sequences were then merged, quality filtered and dereplicated using functions of fastq_mergepairs, fastx_filter and derep_fullength in Vsearch17. All the unique sequences were then clustered at 97% (via cluster_fast) followed by chimera removing (via uchime3_denovo). At last, the non-chimera sequences were re_clustered at 97% to generate OTU representative sequences and an OTU table. Alpha-diversity metrics (Chao1, ACE), and beta diversity metrics (Bray-Curtis dissimilarity) were estimated using the diversity plugin. Taxonomy was assigned to OTUs using the classify-sklearn naive Bayes taxonomy classifier in feature-classifier plugin18 in QIIME 2 against the nifH database formatted for the QIIME2 pipeline originally released as v2.0.5 (2023)19. Non-metric multidimensional scaling (NMDS) based on the Bray-Curtis distance was performed to display community patterns using the “vegan” package20.
Technical Validation
To ensure the generation of unbiased data, random sampling was employed during the collection of wheat root zone samples from each site, targeting plants exhibiting no signs of disease or pest infestation. The extracted DNA was evaluated using a NanoDrop One spectrophotometer, alongside electrophoresis on a 1% agarose gel. The raw amplicon sequencing data underwent a comprehensive quality control pipeline, encompassing the removal of low-quality reads and chimeric sequences.
Code availability
The code is available at the figshare: https://figshare.com/articles/dataset/Root-zone-diazotrophs/2975651022.
References
Collavino, M. M. et al. N2-fixing communities in agricultural soils. Environ Microbiol 16, 3211–3223 (2014).
Timofeeva, A. M., Galyamova, M. R. & Sedykh, S. E. Plant growth-promoting soil bacteria: nitrogen fixation, phosphate solubilization, siderophore production, and other biological activities. Plants. 12, 40–74 (2023).
Tarolli, P., Luo, J., Park, E., Barcaccia, G. & Masin, R. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. iScience 27(2), 108830 (2024).
Kuypers, M. M., Marchant, H. K. & Kartal, B. “The microbial nitrogen-cycling network.”. Nature Reviews Microbiology 16(5), 263 (2018).
Compant, S., Clément, C. & Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biology and Biochemistry 42(5), 669–678 (2010).
Qin, Y., Druzhinina, I. S., Pan, X. & Yuan, Z. Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnology Advances 34(7), 1245–1259 (2016).
Xu, Z. et al. Exogenous application of poly-c-glutamic acid enhances stress defense in Brassica napus L. seedlings by inducing cross-talks between Ca2+, H2O2, brassinolide, and jasmonic acid in leaves. Plant Physiology Biochemistry 118, 460–470 (2017).
Zilaie, M. N., Arani, A. M., Etesami, H., Dinarvand, M. & Dolati, A. Halotolerant plant growth-promoting rhizobacteria-mediated alleviation of salinity and dust stress and improvement of forage yield in the desert halophyte Seidlitzia rosmarinus. Environ. Exp. Bot. 201, 104952 (2022).
Upadhyay, S. K. & Singh, D. P. Effect of salt-tolerant plant growth-promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant biology. 17(1), 288–293 (2015).
Chen, S. et al. Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 7(1), 1–13 (2019).
Haroon, U. et al. Halotolerant plant growth-promoting rhizobacteria induce salinity tolerance in wheat by enhancing the expression of SOS genes. J. Plant Growth Regul. 41, 1–14 (2021).
Tautges, N. E., Sullivan, T. S., Reardon, C. L. & Burke, I. C. Soil microbial diversity and activity linked to crop yield and quality in a dryland organic wheat production system. Applied Soil Ecology 108, 258–268 (2016).
Zhao, H., Zhang, L., Liu, M., Wang, X., Oljira, A. M. Trichoderma Combined with 1-Aminocyclopropane-1-carboxylic acid (ACC) Soil Amendments Modulates the Root Microbiome and Improves Wheat Growth Under Salinity Stress, Plant Stress, https://doi.org/10.1016/j.stress.2025.100785 (2025).
Zhao, H. et al. Metagenomic and network analyses reveal key players in nitrification in upland agricultural soils. Environmental Microbiology 25(11), 2636–2640 (2023).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature biotechnology 37, 852–857 (2019).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal 17, 10–12 (2011).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Bokulich, N. A. et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 1–17 (2018).
Moynihan, M. A. & Reeder, C. F. nifHdada2 GitHub repository, v2.0.5. Zenodo. https://doi.org/10.5281/zenodo.7996213 (2023).
Oksanen, J. et al. vegan: Community Ecology Package. R package version 2.5-6 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP575988 (2025).
Huicheng, Z. Root-zone-diazotrophs. figshare. Dataset. https://doi.org/10.6084/m9.figshare.29756510.v1 (2025).
Acknowledgements
This study was supported by the National Key Research and Development Program of China (2023YFD190260201).
Author information
Authors and Affiliations
Contributions
H.Z. conceived and designed the project. H.Z., M.L. and X.F. formed the experiment, conducted the microbiological analysis, and created the graphs. H.Z. wrote the paper. H.Z., L.Z. and AMO reviewed the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, H., Liu, M., Zhang, L. et al. Root zone diazotrophs of wheat in coastal saline soils from water-scarce regions of the Bohai Sea. Sci Data 12, 1447 (2025). https://doi.org/10.1038/s41597-025-05803-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-025-05803-1