Abstract
Access to high-quality marine geophysical and biogeochemical in-situ data poses a challenge for model evaluation and parameter calibration of the Coastal China Sea (CCS). We describe a new regional ocean database for CCS (RODCCS) with original data from six repositories. The database covers the region of 116–135°E in longitude and 20–42°N in latitude, which embraces the Bohai Sea, the Yellow Sea, the East China Sea and a part of the Sea of Japan. About 3.9 million data points are collected and sorted according to variable types, including temperature, salinity, dissolved oxygen, silicate, nitrate, nitrite, ammonium, phosphate, Chlorophyll a, dissolved inorganic carbon, dissolved organic carbon, and particulate organic carbon. These data are quality-controlled (QCed) with six QC checks and stored in a Network Common Data Format (NetCDF) file. RODCCS includes twelve NetCDF files, each with a unified structure. The database is easily accessed and of high quality after QC checks, making it suitable for a wide range of marine modelling as well as field research for the CCS.
Similar content being viewed by others
Background & Summary
The coastal region is closely tied to human life, featuring intricate ecological and economic impacts. Anthropogenic activities have led to a variety of environmental issues in coastal waters1. Hypoxia (oxygen concentration lower than 2 mg L−1) has frequently been reported in the Coastal China Sea (CCS) over recent decades2,3,4, driven by both anthropogenic activities and climate change5,6,7. The Yangtze River estuary is the largest estuary in the CCS. In its adjacent coastal ocean, shallow hypoxia has been observed in summer and autumn in recent years8,9,10. It has a significantly negative impact on environmental health and subsequently on ecological community compositions and fisheries11,12. Therefore, a sufficient understanding of their underlying mechanisms and exploring solutions to alleviating hypoxia are very urgent4,13. Continental shelves absorb atmospheric CO2 at a rate of about 0.2 Pg C yr−1, accounting for approximately 13%–15% of the current global oceanic CO2 uptake14,15,16. The CCS, one of the largest continental shelves on Earth17, is considered a region with significant carbon sink potential18,19,20 and deserves great effort in quantifying its carbon fluxes and predicting its response to future climate change.
To grasp the physicochemical characteristics of coastal waters, researchers use marine physical coupled biogeochemical models, which serve as crucial tools in testing hypotheses and quantifying fluxes of elemental cycles. Model evaluation and parameter calibration require a large volume of observational data. However, accessing these data poses a challenge due to their dispersed distribution. Furthermore, a strict quality control (QC) for different types of in-situ observational data is necessary.
Considering the recently recognised importance of CCS in the carbon cycle and the increased number of reported hypoxia events, a regional ocean database for CCS (RODCCS) is compiled in this study to offer comprehensive and reliable observational data for both modelling and field research21. RODCCS includes data from six repositories, one of which comprises unpublished data from authors (Table 1). Our database covers the region of 116° to 135° E in longitude and 20° to 42° N in latitude. It encompasses the Bohai Sea, the Yellow Sea, and the East China Sea, as well as a part of the Sea of Japan and its sampling depths span from the surface to 6984 meters (Fig. 1a,b). The sampling years of the data span from 1985 to 2021 (Fig. 1b). The database includes twelve variables, i.e., temperature (4,348,536 data points), salinity (4,325,295 data points), and dissolved oxygen (DO, 4,235,725 data points), silicate (726,086 data points), nitrate (745,908 data points), nitrite (246,347 data points), ammonium (29,526 data points), phosphate (729,507 data points), Chlorophyll a (Chl a, 73,715 data points), dissolved inorganic carbon (DIC, 128,744 data points), dissolved organic carbon (DOC, 196,919 data points), particulate organic carbon (POC, 25,190 data points) concentrations.
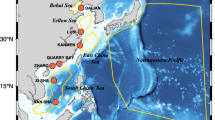
Spatio-temporal distribution of RODCCS. (a) Blue, black, green, purple, pink and yellow points represent data from Argo, NESSDC, GLODAPv2, CCHDO, CoastDOM and R2R, respectively. (b) Hovmoller diagram of spatiotemporal distribution of RODCCS. The shadings indicate the log10-transformed numbers of data points. The white shading indicates the absence of data. The orange line indicates the log10-transformed numbers of data points for each year during the period from 1985 to 2021.
We apply six strict QC checks to each variable of the collected in-situ data. The QC includes location check, depth check, constant value check, value range check, vertical gradient check and time reversal check (Table 2). After QC, irrelevant, failed and acceptable data are marked with flags of 1, 2 and 3, respectively (Table 3). The numbers of failed data by the six QC checks for the twelve variables are presented in Table 4. We store the data in twelve NetCDF format files with one variable in each file. There is a uniform structure in each file where longitude, latitude, depth, sampling time, data source ID, QC flag and variable values are included. It provides the spatial and temporal attributes, original repository information, QC check result, and value of each data point. RODCCS provides quality-controlled observational data for model evaluation as well as inter-comparisons of different databases, making it suitable for a wide range of marine modelling and field research.
Methods
The original data of RODCCS are from six repositories, encompassing observational data in various formats, including Comma-Separated Values (.csv), NetCDF, Excel Open XML Spreadsheet (.xlsx), Copy Number Variation (.cnv), and Tab-Separated Values (.tab). The procedures of RODCCS compilation are summarised in Fig. 2.
Array for real-time geostrophic oceanography (Argo) is an international program that measures water properties across the world’s ocean using a fleet of robotic instruments that drift with the ocean currents and move up and down between the surface and a mid-water level22. On the top of every Argo float is a conductivity, temperature, pressure sensor which measures temperature within an accuracy of 0.001 °C and pressure within 0.1 dbar, and calculates salinity using conductivity, temperature, and pressure within 0.001 psu (practical salinity units). Biogeochemical-Argo (BGC-Argo) is the extension of the Argo array of profiling floats to include floats that are equipped with biogeochemical sensors for pH, oxygen, Chl a, nitrate concentrations, suspended particles, and downwelling irradiance. On the Euro-Argo European Research Infrastructure Consortium (ERIC) website (https://fleetmonitoring.euro-argo.eu)23, we select data from the China Sea Institute of Oceanology (CSIO), Korea Meteorological Administration (KMA), and Korea Ocean Research & Development Institute (KORDI) data centre for download. They all fall under the category of BGC-Argo data. All data from CSIO, KMA, and KORDI are exclusively adjusted data, which are raw sensor outputs and remain institutionally archived. These Argo data have undergone algorithmic processing and environmental compensation procedures24. CSIO, KMA and KORDI provide depth data rather than sensor-measured pressure data, which are depth values derived directly from pressure sensors and synthetically reconstructed layers from multi-sensor fusion25. CSIO delivers both real-time (automatically quality-controlled) and delayed-mode (expert-validated) adjusted data. KMA and KORDI provide real-time adjusted data without delayed-mode products due to shortened float deployments, which are transmitted via satellite in near real-time. Delayed-mode data undergo rigorous quality control protocols, including sensor calibration, salinity bias adjustment, and outlier removal25,26. For BGC-Argo, delayed-mode processing further integrates laboratory analytical validation25. Real-time QC of Argo detects physically implausible values and ensures vertical profile consistency, while delayed-mode QC combines expert manual verification with regional climatological datasets to identify biases24. We obtain original data in NetCDF files that include sampling sites, sampling times, DO concentration, salinity and temperature. We then use MATLAB to extract these data. The depth of each data point is converted from pressure using the seawater toolbox in MATLAB.
Climate and Ocean Variability, Predictability and Change (CLIVAR) and Carbon Hydrographic Data Office (CCHDO) support oceanographic research by providing access to high quality, global, vessel-based Conductivity, Temperature and Depth (CTD) and hydrographic data from Global Ocean Ship (GO-SHIP), World Ocean Circulation Experiment (WOCE), CLIVAR and other repeat hydrography programs27,28. The electrochemical Sea-Bird SBE43 sensor is utilised to measure DO concentration in CCHDO29. Data are retrieved from the CCHDO database through its advanced search platform (https://cchdo.ucsd.edu/search/advanced) in.xlsx format. Ten variables, i.e., Chl a, DIC, DO, DOC, nitrate, nitrite, phosphate, and silicate concentrations, salinity, and temperature, are obtained from the CCHDO database. The depth of each data point is also converted from pressure using the seawater toolbox in MATLAB.
We retrieve four datasets from the National Earth System Science Data Center (NESSDC, http://www.geodata.cn/) and integrate them into our database. We conduct a targeted search for cruise expeditions along the coastal regions of China, with a specific emphasis on Chl a concentration. Following the submission of a formal request and subsequent grant of access by the website administrators, we obtain the Yellow Sea and East China Sea Chl a concentration data for 2011–2013, the Bohai Sea Chl a concentration data for 2015 and 2017, and the China Coastal Chl a concentration data for 2009–2012 (offshore CTD Chl a concentration measurements in CCS), in four.xlsx format files respectively. The in-situ Chl a concentration data from NESSDC are unpublished in the scientific literatures. The data authors are authors of this study, and permission to use the data has been granted. The Chl a concentrations in these datasets are all determined using the Trilogy fluorometer technique. We extract Chl a concentration data along with their corresponding sampling location and time information.
The Coast Dissolved Organic Matter (CoastDOM) database includes comprehensive coastal DOM concentration data in a single repository, making it openly and freely available to different research communities30. In CoastDOM, the concentrations of 81% samples are determined using a High-Temperature Catalytic Oxidation (HTCO) analyser for DOC concentration, with the remaining 19% determined by a combination of wet chemical oxidation (WCO) and/or UV digestion. Data from the CoastDOM are downloaded from the website (https://doi.pangaea.de/10.1594/PANGAEA.964012) in.tab format. Five variables, i.e., Chl a, DIC, DOC, POC, and ammonium concentrations, together with their corresponding sampling location and time information, are extracted with MATLAB.
Global Ocean Data Analysis Project Version 2 (GLODAPv2) is a synthesis activity for ocean surface to bottom biogeochemical data collected through chemical analysis of water samples31,32,33. GLODAP deals only with bottle data and CTD data at bottle trip depths. The consistency of its data product is estimated to be better than 0.005 for salinity, 1% for oxygen, 2% for nutrients, 4 μmol/kg for DIC concentration and total alkalinity, and 0.01–0.02 for pH, indicating a high level of precision and reliability across these measurements. We download data from the Pacific Ocean part of GLODAPv2.2023 (released in 2023) via the GLODAPv2 portal (https://glodap.info/index.php/merged-and-adjusted-data-product-v2-2023/) in a.csv format file34. We extract nine variables, i.e., Chl a, DO, DOC, nitrate, nitrite, phosphate, silicate concentrations, salinity and temperature, together with their corresponding sampling location and time information.
The Rolling Deck to Repository (R2R), with their global capability and diverse array of sensors and research vessels, is an essential mobile observing platform for ocean science35,36. Temperature and salinity are measured with a CTD profiler, and DO concentration is measured by an oxygen sensor. R2R provides essential documentation and standard products for each expedition, as well as tools to document shipboard data acquisition activities while underway. Data collected on every expedition are of high value, given the high cost and increasingly limited resources for ocean exploration. We download the cruise data from the Pacific Ocean on the R2R website (https://www.rvdata.us/search?keyword=ctd&zoom=1&x=0&y=2646652.0332176173&projection=M). Twenty-three cruise files in.cnv format from this repository within the region of RODCCS are selected for further analysis. Ultimately, we extract the values for DO concentration, salinity and temperature, and the location and temporal information of each data point, and then include them in RODCCS.
In order to control the quality of the in-situ data, we apply six types of checks for each variable. The QC includes location check, depth check, constant value check, value range check, vertical gradient check and time reversal check. The six QC checks are listed in Table 2 and explained below, and data points are flagged with 1, 2, or 3 when they are irrelevant, have failed or passed the specific check (Table 3). Flags of each data point are saved in the NetCDF format files for all variables.
Location check
The location check ensures the accuracy of the sampling locations within the CCS region defined in this study37,38. Due to the diversity of data sources and the varying sampling locations across different cruises, the collected dataset includes data outside the CCS region. Data points outside the study area fail this inspection and are not subject to the subsequent five inspections, while only data points that passed undergo the subsequent inspections.
Depth check
The depth check assesses whether the sampling depth is shallower than the corresponding seabed depth estimated with the General Bathymetric Chart of the Oceans (GEBCO) data37,38,39. Data points either above sea level or deeper than the seabed fail this check and are not applied in the subsequent QC checks (Fig. 3).
Results of the depth check of temperature, salinity, DO, silicate, nitrate, nitrite, ammonium, phosphate, Chl a, DIC, DOC, and POC concentrations (a–l). Black lines are the seabed topography at 123°E (a–i) and 23.08°N (j–l) estimated from the GEBCO dataset. Black, blue and red circles are original data, data passed the QC and failed the QC (data below the seabed or above the sea level), respectively.
Constant value check
This check identifies consecutive identical values in the vertical profile, which suggests instrument malfunction or data corruption38, resulting in sampling values that do not change with variations in depth or time. We analyse the data distributions of each profile and find that anomalous values typically occur three or more times consecutively. As a result, data points with occurrences of more than three consecutive identical values in a vertical profile are identified as failed data and not applied in the subsequent checks.
Value range check
For variables that conform to or approximate a normal distribution (log-transformed DOC, Chl a, ammonium concentrations, and salinity), we employ the Chauvenet’s criterion to identify outliers. For a dataset comprising N measurements, any value that deviates from the mean with a probability of less than 1/(2 N) is classified as a suspicious outlier40,41. We determine the critical value using MATLAB’s norminv function, which requires the mean (mu) and standard deviation (sigma) of the data (Eq. 1). Given that this method is a two-sided test and only data at the tail with high values will be identified, the threshold is calculated with 1-1/(4 N)42,43 (Eq. 1). Any measurements exceeding the critical value are taken as outliers, with the remaining measurements as pass values (Fig. 4).
Where mu and sigma are the mean and standard deviation, respectively.
Results of the value range check of salinity, Chl a, DOC and ammonium concentrations, temperature, DO, silicate, phosphate, nitrate, nitrite, DIC, and POC concentrations (a–l). Black, blue and red bars represent numbers of original, passed and failed data, respectively. Black dashed lines and blue solid lines represent the fitting-lines of the data distribution before and after QC, respectively (a–d). Red dashed lines indicate Chauvenet’s critrion thresholds (a–d) and magenta dashed lines indicate IQR thresholds (e–l), respectively. Subfigures are used to better show the distributions of failed data, with the y-axis values in each subfigure representing the quantities of the respective data points (a,e,f,j).
Log-transformed DOC, Chl a and ammonium concentrations, and salinity undergo the value range check with the Chauvenet’s criterion method. Since Chl a, ammonium, and DOC concentrations don’t have negative values and may have very low values, the threshold for these variables is set between 0 and the Critical value defined by Eq. 1. We also compare these thresholds with the ranges collected from the literatures (Table 1), to validate the rationality of the thresholds applied in this check.
For variables that do not follow a normal or approximately normal distribution (temperature, and DO, silicate, nitrate, nitrite, phosphate, DIC, POC concentrations), we utilise the Interquartile Range (IQR) method for outlier identification, as shown in Eqs. 2–444. Iqr, defined as the difference between the 75% quantile and 25% quantile of a variable, together with published value ranges45,46,47, is applied in determining the upper (Upper Bound) and lower (Lower Bound) bounds for outlier identification (Eqs. 3, 4). Through comparative analysis of literature-reported ranges (Table 1) and bounds derived from varying Iqr coefficients, a coefficient of 2 is ultimately determined in Eqs. 3, 4, which produces better consistency between both ranges. Data points outside the calculated ranges are marked as outliers (Fig. 4).
Where Q1 and Q2 are the 25% quartile and 75% quartile of a variable, respectively.
Vertical gradient check
This check is conducted to identify excessive decreases or increases in variable values over a depth range. A gradient is defined as:
Where \({\nu }_{{\rm{i}}}\) and \({\nu }_{{\rm{i}}-1}\) are the values of a variable at the current depth level and the previous depth level (shallower). zi and zi-1 are the depths (in meter) of the current depth level and the previous depth level (shallower), respectively. k is the number of data points in a vertical profile.
We firstly sort the data points with identical latitude and longitude coordinates into identical vertical profiles. We analyse the number of data points per vertical profile and find that most sampling events contain over ten data points. If the number of samples in a particular vertical profile exceed or is equivalent to ten, a vertical gradient check is conducted. Otherwise, all data points in a vertical profile are marked as irrelevant. Since data distributions of nitrite, ammonium, Chl a, DIC, DOC, and POC concentrations are highly dispersive and often do not meet this criterion, vertical gradient check is only performed on the six variables of temperature, salinity, DO, silicate, nitrate, and phosphate concentrations.
For each vertical profile, a surface-to-bottom check sequence is adopted. The value of the shallowest sampling point is validated against the World Ocean Atlas 2013 (WOA13) annual climatological data48. If the value of a data point is out of the value estimated from WOA13 at its corresponding location by ± n%, it is flagged as an outlier. Then the data point of the next deeper level is estimated until a value within the acceptable range is identified, which is then adopted as the first sampling point. The spatial distributions of salinity and DO concentration are significantly different from the other four variables; therefore, we set different n values to ensure that our selection of the first sampling point is appropriate. We determine the values of n by comprehensively analysing the results obtained from different initial sampling points, which are generated by varying n. The n values resulting in accurate outlier identification are selected. Finally, the n values used in this check for salinity, DO concentration, and the other four variables are 20, 40, and 100, respectively. The next sampling point (νi) at depth zi and the starting point (νi−1) at depth zi−1 are selected. If the depth interval Δz (|zi - zi-1|) is greater than 10 meters, a vertical gradient check is performed. Analysis of gradients in vertical profiles indicates that smaller Δz values (Δz < 10 m) reduce the value of the denominator in Eq. 5, resulting in unreasonably large gradients. Numerous valid data points are misidentified as outliers. Conversely, if Δz is less than 10 meters, the search moves upward to a shallower point that has passed the vertical gradient check as the i−1 point. Data points within 10 meters vertically from the starting point are irrelevant for this check. To better represent the differences in gradient ranges between surface and deep waters (e.g. due to physical or biogeochemical influences), every data point has been categorised into the shallow water group (depth ≤ 400 m) or the deep water group (depth > 400 m). Data point with gradients exceeding the maximum gradient value (MGV) fails this check and are flagged (Fig. 5). We compare the results with different MGV values and verify their corresponding locations and values of outliers. The MGV values with which obvious outliers are identified are applied in this check (Table 5).
Vertical gradient check results of temperature, salinity, DO, silicate, nitrate, phosphate concentrations. Black, blue, red and yellow dots represent original, passed, failed, and irrelevant data, respectively (a,b,e,f,i,j,m,n,q,r,u,v). Blue, red and yellow circles represent passed, failed, and irrelevant data points in randomly selected vertical profiles (c,g,k,o,s,w) and their corresponding vertical gradient respectively (d,h,l,p,t,x). Red dashed lines indicate MGVs as described in Table 5.
Time reversal check
This check identifies instances where data points are recorded out of temporal sequence, leading to misinterpretations of temporal trends. Within the same sampling event, data points that do not conform to an increasing chronological order are flagged as failed data. Data points contain sampling time information of only year and month, but without day, hour, and minute, are marked as irrelevant, as they do not provide sufficient temporal resolutions required for this check37.
Evaluation of QCs for RODCCS
After QC, we employ the dichotomous metrics of True Positive Rate (TPR), False Positive Rate (FPR), and True Negative Rate (TNR) to evaluate QC performance (Fig. 2). TPR reflects the QC’s ability to correctly retain valid data points. FPR indicates the proportion of normal data erroneously flagged as anomalies. TNR measures the specificity in preserving true negative instances38,49. These metrics quantify the trade-off between detection efficacy and error control, thereby providing a comprehensive evaluation of the discriminative capacity of the QC system. Optimal performance of QC is achieved when both TPR and TNR are maximised, and FPR is minimised38. These dichotomous metrics are initially proposed by Yerushalmy and defined as follows38,49:
In these equations, NTP, NFN, NFP and NTN represent the numbers of true positives, false negatives, false positives, and true negatives, respectively. For measurement, the benchmark dataset provides the true passed or rejected flags, which are then compared against the QC results (pass or fail). We use GLODAPv2 as a benchmark dataset to evaluate the performance of our QC of RODCCS due to its comprehensive quality control procedures for multiple variables. Given that the GLODAPv2 dataset contains quality check indicators only for salinity, DO, silicate, nitrate, and phosphate concentrations, the evaluation of QC checks for RODCCS is conducted exclusively on these five variables.
Data Records
RODCCS is stored in twelve NetCDF format files, and each file encompasses ten variables, with nine common foundational information variables and one unique variable. The variable descriptors in the RODCCS_temperature.nc file are listed below, and the other files in RODCCS follow the same format to organise variables in each record:
Variables:
Longitude
Size: 4348536×1
Dimensions: data_number
Datatype: double
Attributes:
standard_name = ‘longitude’
units = ‘degrees_east’
FillValue = NaN
valid_min = −180
valid_max = 179.9984
variable properties = ‘common foundational information variable’
Latitude
Size: 4348536 × 1
Dimensions: data_number
Datatype: double
Attributes:
standard_name = ‘latitude’
units = ‘degrees_north’
FillValue = NaN
valid_min = −78.643
valid_max = 89.9909
variable properties = ‘common foundational information variable’
Depth
Size: 4348536 × 1
Dimensions: data_number
Datatype: double
Attributes:
standard_name = ‘depth’
units = ‘m’
FillValue = NaN
valid_min = −4.639
valid_max = 61228.213
variable properties = ‘common foundational information variable’
Year
Size: 4348536 × 1
Dimensions: data_number
Datatype: double
Attributes:
standard_name = ‘year’
units = ‘years’
FillValue = NaN
valid_min = 1978
valid_max = 2022
variable properties = ‘common foundational information variable’
Month
Size: 4348536 × 1
Dimensions: data_number
Datatype: double
Attributes:
standard_name = ‘month’
units = ‘months’
FillValue = NaN
valid_min = 1
valid_max = 12
variable properties = ‘common foundational information variable’
Day
Size: 4348536 × 1
Dimensions: data_number
Datatype: double
Attributes:
standard_name = ‘day’
units = ‘days’
FillValue = NaN
valid_min = 1
valid_max = 31
variable properties = ‘common foundational information variable’
Time
Size: 4348536 × 1
Dimensions: data_number
Datatype: double
Attributes:
standard_name = ‘sampling time’
units = ‘minute’
FillValue = NaN
valid_min = 0
valid_max = 2400
variable properties = ‘common foundational information variable’
QC flag
Size: 4348536 × 1
Dimensions: data_number
Datatype: double
Attributes:
standard_name = ‘quality control of sampling data point’
units = ‘xxxxxx, x equals 1 or 2 or 3’
FillValue = NaN
valid_min = 211111
valid_max = 333333
variable properties = ‘common foundational information variable’
Data Source ID
Size: 4348536 × 1
Dimensions: data_number
Datatype: double
Attributes:
standard_name = ‘Source of data point’
units = ‘constant’
FillValue = NaN
valid_min = 1
valid_max = 6
variable properties = ‘common foundational information variable’
Temperature
Size: 4348536 × 1
Dimensions: data_number
Datatype: double
Attributes:
standard_name = ‘Temperature of seawater’
units = ‘°C’
FillValue = NaN
valid_min = −57.5261
valid_max = 99
variable properties = ‘unique variable’
In the attribute description of each NetCDF file, a comprehensive summary is provided to introduce the sources of in-situ data, where each point is distinctly recognised by a unique source identifier (Data Source ID). The Data Source ID values are consecutive integers from 1 to 6, denoting the six data repositories of Argo, CCHDO, NESSDC, CoastDOM, GLODAPv2 and R2R, respectively (Table 1). The QC flag is a six-digit integer, with each digit (x) representing the outcome of a QC check (Table 3). Longitude, Latitude and Depth serve as the location descriptors of the data point. Year, Month, Day and Time serve as the time descriptors of each data point. The twelve files of RODCCS in NetCDF format can be accessed on Figshare21 using the link (https://doi.org/10.6084/m9.figshare.28532210). ‘NaN’ denotes missing data.
Technical Validation
The six QC checks have significantly improved the data quality, and the number of data failed during all checks are shown in Table 4.
Location check
The number of data points excluded by the location check are the most abundant (Table 4). Among the twelve variables, the largest number of data points that failed the check is observed in temperature, with 1,496,903 data points. Conversely, the lowest number of failed data points is recorded for POC concentration, with 24,996 data points. DIC concentration has the highest percentage of failed data points relative to its total number of data points, accounting for 99.42%. In contrast, the lowest percentage is found in DO concentration, which is 32.21%.
Depth check
During the depth check, anomalies above sea level are exclusively found in temperature, salinity, and DO concentration. The remaining outliers exceed the maximum sea depth estimated with the GEBCO data, predominantly occurring at several deepest points of each single vertical sampling event. Given the dispersive distribution of data points, to more effectively demonstrate the effectiveness of the depth check, we select the vertical sections at 123°E and 23.08°N of the domain, which have a higher number of data points. The spatial distribution of outliers in seawater temperature, salinity, and DO concentration exhibits a high degree of similarity, indicating a strong likelihood that these outliers are derived from the same sampling event (Fig. 3a–c). A similar pattern is also observed for the concentrations of nitrate, nitrite, and phosphate (Fig. 3e,f,h). No significant outliers are detected in the concentrations of silicate, DIC, DOC, and POC (Fig. 3d,g–l) in the selected transect.
Constant value check
The disparity in the number of outliers identified by constant value check across different variables is substantial. A total of 89,004 and 232,935 outliers are identified for temperature and salinity, which account for 2.05% and 5.39% of the original data, respectively. In contrast, no outliers are identified for DIC and POC concentrations.
Value range check
In the value range check, outliers for salinity are much less than for the remaining variables. Figure 4a–d present the outcomes of the Chauvenet’s criterion. Figure 4a shows both unusually low salinity values (values below 23.71 psu), while Fig. 4c shows excessively high value for DOC concentration (values outside the range of 0 to 371.53 µmol/L). Chl a and ammonium (Fig. 4b,d) concentrations do not yield any outliers in this check, indicating that the sampled values for these variables are within a reasonable range. Analysis of the original and passed fitting-line derived from the salinity data distribution reveals that the distribution conforms more closely to a normal distribution after eliminating outliers (Fig. 4a).
Figure 4e–k demonstrates the effectiveness of the IQR method. Figure 4e shows that the IQR method successfully identifies excessively high temperature values (e.g., values greater than 42.07 °C), and Fig. 4f,j,l show the unusually high DO, phosphate, POC concentrations (e.g., values greater than 338.56, 7.4, 899.75 µmol/L) are identified.
Vertical gradient check
The vertical gradient check identifies values that exhibit substantial discrepancies from values of their adjacent data points. The highest number of data points of 85,526, which failed the check, is observed in DO concentration (Table 4). And DO concentration also has the highest percentage of failed data points relative to the total number of data points, accounting for 2.02%.
Irrelevant values of temperature, salinity, and DO concentration are predominantly distributed in shallow waters (Fig. 5b,f,j). This is attributed to that sampling events for these three variables in shallow waters often have fewer sampling points (less than 10), and thus cannot be regarded as continuous vertical profiles in this check. From randomly selected temperature and salinity sampling events, it can be observed that the vertical gradient check accurately identified the anomalous values in a set of sampling data (Fig. 5c,d,g,h).
Anomalous values of DO concentration are mostly located in shallow water and are often among the first few points of a single sampling event (Fig. 5j). This can be attributed to measurement errors that occur when the sampling instrument is initially activated. For nutrient concentrations, our vertical gradient check successfully identifies anomalously low values in the deep sea (Fig. 5n,r,v) and high values in the shallow sea (Fig. 5r), thereby rendering the vertical distribution of the data more reasonable (Fig. 5m,n,q,r,u,v).
Time reversal check
This check results in the identification of a modest number of outliers, and the highest number of data points that failed the check is observed in temperature, with 27,232 data points (Table 4), while the highest percentage of failed data points relative to the total number of data points is observed for phosphate concentration, accounting for 2.98%. It is important to note that no outliers are identified by this check for ammonium, DIC, DOC, and POC concentrations (Table 4). Given that the majority of the sampling points are not time-series data, therefore, most of the data are marked as irrelevant in this check.
Evaluation of QCs for RODCCS
Table 6 presents a comprehensive assessment of QC checks for five variables. Overall, the QC checks demonstrate excellent performance, with high TPR and TNR values for the five variables. This indicates a robust capacity to accurately identify true positives and true negatives. The value range checks achieve a TNR as much as 100% for all variables, i.e., the outliers identified by our value range check method are identical with the outliers identified by the default QC check for GLODAPv2, suggesting high efficacy of the value range check employed.
Code availability
The source codes for data extraction, QC checks, writing data into NetCDF files, and data validation and visualisation used in compiling RODCCS are written in MATLAB and are available at https://github.com/BGM-USD2020/RODCCS_codes.git.
References
Xiong, L. et al. Nutrient input estimation and reduction strategies related to land use and landscape pattern (LULP) in a near-eutrophic coastal bay with a small watershed in the South China sea. Ocean Coastal Management. 206, 105573 (2021).
Zhu, Z.-Y. et al. Hypoxia off the Changjiang (Yangtze River) estuary and in the adjacent East China Sea: Quantitative approaches to estimating the tidal impact and nutrient regeneration. Marine Pollution Bulletin. 125, 103–114 (2017).
Zhou, F. et al. Investigation of hypoxia off the Changjiang Estuary using a coupled model of ROMS-CoSiNE. Progress In Oceanography. 159, 237–254 (2017).
Zhou, F. et al. Coupling and decoupling of high biomass phytoplankton production and hypoxia in a highly dynamic coastal system: the Changjiang (Yangtze River) Estuary. Frontiers in Marine Science. 7, 259 (2020).
Zhang, H. et al. A numerical model study of the main factors contributing to hypoxia and its interannual and short-term variability in the East China Sea. Biogeosciences. 17, 5745–5761 (2020).
Howarth, R. et al. Coupled biogeochemical cycles: eutrophication and hypoxia in temperate estuaries and coastal marine ecosystems. Frontiers in Ecology the Environment. 9, 18–26 (2011).
Qun, D. Z. Zhenya and ecology. Analysis on environmental pollution in china’s coastal ecosystem. Journal of Resources and Ecology. 10, 424–431 (2019).
Zhang, J. et al. Editorial: Eutrophication and hypoxia and their impacts on the ecosystem of the Changjiang Estuary and adjacent coastal environment. JOURNAL OF MARINE SYSTEMS. 154, 1–4 (2016).
Zhou, F. et al. Recent progress on the studies of the physical mechanisms of hypoxia off the Changjiang(Yangtze River)Estuary. Journal of Marine Sciences. 39, 22–38 (2021).
Li, X. A. et al. Nitrogen and phosphorus budgets of the Changjiang River estuary. Chinese Journal of Oceanology and Limnology. 29, 762–774 (2011).
Liu, F. et al. Marine environmental pollution, aquatic products trade and marine fishery Economy—An empirical analysis based on simultaneous equation model. Ocean & Coastal Management. 222, 106096 (2022).
Xu, W. & Zhang, Z. Impact of coastal urbanization on marine pollution: Evidence from China. International Journal of Environmental Research Public Health. 19, 10718 (2022).
Ding, X. et al. Interannual variations in the nutrient cycle in the central Bohai Sea in response to anthropogenic inputs. Chemosphere. 313, 137620 (2023).
Poloczanska, E. The IPCC Special Report on the Ocean and Cryosphere in a Changing Climate. 2018 Ocean Sciences Meeting., (2018).
Borges, A. V., Delille, B. & Frankignoulle, M. Budgeting sinks and sources of CO2 in the coastal ocean: Diversity of ecosystems counts. Geophysical research letters. 32, (2005).
Dai, M. et al. Carbon Fluxes in the Coastal Ocean: Synthesis, Boundary Processes, and Future Trends. Annual Review of Earth and Planetary Sciences. 50, 593–626 (2022).
Na, R. et al. Air-sea CO2 fluxes and cross-shelf exchange of inorganic carbon in the East China Sea from a coupled physical-biogeochemical model. Science of The Total Environment. 906, 167572 (2024).
Zhai, W. & Dai, M. On the seasonal variation of air-sea CO2 fluxes in the outer Changjiang (Yangtze River) Estuary, East China Sea. Marine Chemistry. 117, 2–10 (2009).
Tseng, C.-M., Shen, P.-Y. & Liu, K.-K. Synthesis of observed air–sea CO 2 exchange fluxes in the river-dominated East China Sea and improved estimates of annual and seasonal net mean fluxes. Biogeosciences. 11, 3855–3870 (2014).
Tseng, C. M. et al. CO2 uptake in the East China Sea relying on Changjiang runoff is prone to change. Geophysical Research Letters. 38, (2011).
Wang, C. et al. A regional ocean database for the Coastal China Sea. Figshare https://doi.org/10.6084/m9.figshare.28532210 (2025).
Wang, X.-l, Qin, B. & Liu, P.-s Argo data sharing system based on grid technology. Computer Engineering and Design. 30, 3634–3637 (2009).
Euro-Argo: Argo Fleet Monitoring– Euro-Argo, available at: https://fleetmonitoring.euro-argo.eu/, last access: (2022).
Li, Z. Q., Liu, Z. H. & Lu, S. L. Global Argo data fast receiving and post-quality-control system. IOP Conference Series: Earth and Environmental Science. 502, 012012 (2020).
Chai, F. et al. Monitoring ocean biogeochemistry with autonomous platforms. Nature Reviews Earth & Environment. 1, 315–326 (2020).
Guo, M. et al. Efficient biological carbon export to the mesopelagic ocean induced by submesoscale fronts. Nature Communications. 15, 580 (2024).
Swift, J. Operation of the CLIVAR & Carbon Hydrographic Data Office at UCSD/SIO. National Science Foundation. (2003).
CCHDO Hydrographic Data Office. CCHDO Hydrographic Data Archive, Version 2023-07-24. In CCHDO Hydrographic Data Archive. UC San Diego Library Digital Collections. https://doi.org/10.1016/j.marchem.2010.09.004.
Pensieri, S. et al. Methods and Best Practice to Intercompare Dissolved Oxygen Sensors and Fluorometers/Turbidimeters for Oceanographic Applications. Sensors. 16, 702 (2016).
Lønborg, C. et al. A global database of dissolved organic matter (DOM) concentration measurements in coastal waters (CoastDOM v1). Earth System Science Data. 16, 1107–1119 (2024).
Olsen, A. et al. The Global Ocean Data Analysis Project version 2 (GLODAPv2) - an internally consistent data product for the world ocean. Earth System Science Data. 8, 297–323 (2016).
Olsen, A. et al. An updated version of the global interior ocean biogeochemical data product, GLODAPv2.2020. Earth System Science Data. 12, 3653–3678 (2020).
Lauvset, S. K. et al. An updated version of the global interior ocean biogeochemical data product, GLODAPv2. 2021. Earth System Science Data. 13, 5565–5589 (2021).
Lauvset, S. K. et al. The annual update GLODAPv2. 2023: the global interior ocean biogeochemical data product. Earth System Science Data. 16, 2047–2072 (2024).
Carbotte, S. M. et al. Rolling Deck to Repository: Supporting the marine science community with data management services from academic research expeditions. Frontiers in Marine Science. 9, 1012756 (2022).
Stocks,K. et al. Rolling Deck to Repository (R2R) Perspectives from a Decade of Ocean Data Management. Authorea Preprints. (2022).
Yuan, Y. et al. Design, construction, and application of a regional ocean database: A case study in Jiaozhou Bay, China. Limnology Oceanography: Methods. 17, 210–222 (2019).
Tan, Z. et al. A new automatic quality control system for ocean profile observations and impact on ocean warming estimate. Deep Sea Research Part I: Oceanographic Research Papers. 194, 103961 (2023).
Mayer, L. et al. The Nippon Foundation—GEBCO seabed 2030 project: The quest to see the world’s oceans completely mapped by 2030. Geosciences. 8, 63 (2018).
Limb, B. J. et al. The Inefficacy of Chauvenet’s Criterion for Elimination of Data Points. Journal of Fluids Engineering. 139, 054501 (2017).
Zihao, Z., Lingli, Z. & Yuanqing, W. Outlier removal based on Chauvenet’s criterion and dense disparity refinement using least square support vector machine. Journal of Electronic Imaging. 28, 023028–023028 (2019).
Su, B. et al. A dataset of global ocean alkaline phosphatase activity. Scientific Data. 10, 205 (2023).
Glover, D. M. D., Jenkins, W. J. & Doney, S. C. Modeling Methods for Marine Science. Cambridge University Press. (2011).
Wan, X. et al. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC medical research methodology. 14, 1–13 (2014).
Song, J. et al. Carbon sinks/sources in the Yellow and East China Seas Air-sea interface exchange, dissolution in seawater, and burial in sediments. Science China Earth Sciences. 61, 11 (2018).
Zhao, L. et al. Spatial and vertical distribution of radiocesium in seawater of the East China Sea. Marine pollution bulletin. 128, 361–368 (2018).
Hutchins, D. A. & Capone, D. G. The marine nitrogen cycle: new developments and global change. Nature Reviews Microbiology. 20, 401–414 (2022).
Boyer, T. P. et al. World ocean atlas., (2018).
Yerushalmy, J. Statistical problems in assessing methods of medical diagnosis, with special reference to X-ray techniques. Public Health Reports. 1432–1449 (1947).
Guo, S. et al. Seasonal variation in the phytoplankton community of a continental-shelf sea: the East China Sea. Marine Ecology Progress Series. 516, 103–126 (2014).
Chen, C.-C. et al. Reoxygenation of the hypoxia in the east China sea: A ventilation opening for marine life. Frontiers in Marine Science. 8, 787808 (2022).
Sun, M.-S. et al. Dissolved methane in the East China Sea: distribution, seasonal variation and emission. Marine Chemistry. 202, 12–26 (2018).
Qi, J. et al. Analysis of seasonal variation of water masses in East China Sea. Chinese Journal of Oceanology Limnology. 32, 958–971 (2014).
Shim, J. et al. Seasonal variations in pCO2 and its controlling factors in surface seawater of the northern East China Sea. Continental Shelf Research. 27, 2623–2636 (2007).
Qu, B. et al. Carbon chemistry in the mainstream of Kuroshio current in eastern Taiwan and its transport of carbon into the east China sea shelf. Sustainability. 10, 791 (2018).
Umezawa, Y. et al. Seasonal shifts in the contributions of the Changjiang River and the Kuroshio Current to nitrate dynamics in the continental shelf of the northern East China Sea based on a nitrate dual isotopic composition approach. Biogeosciences. 11, 1297–1317 (2014).
Chou, W.-C. et al. Seasonality of CO 2 in coastal oceans altered by increasing anthropogenic nutrient delivery from large rivers: evidence from the Changjiang–East China sea system. Biogeosciences. 10, 3889–3899 (2013).
Wang, J. et al. Denitrification and anammox: Understanding nitrogen loss from Yangtze Estuary to the east China sea (ECS). Environmental pollution. 252, 1659–1670 (2019).
Li, H. M. et al. Changes in concentrations of oxygen, dissolved nitrogen, phosphate, and silicate in the southern Yellow Sea, 1980–2012: Sources and seaward gradients. Estuarine Coastal. 163, 44–55 (2015).
Gao, L. et al. Nutrient dynamics across the river‐sea interface in the C hangjiang (Y angtze R iver) estuary—E ast C hina S ea region. Limnology Oceanography. 60, 2207–2221 (2015).
Kim, D., Shim, J. & Yoo, S. Seasonal variations in nutrients and chlorophyll-a concentrations in the northern East China Sea. Ocean Science Journal. 41, 125–137 (2006).
Shim, M. J. & Yoon, Y. Y. Long-term variation of nitrate in the East Sea, Korea. Environmental Monitoring Assessment. 193, 1–13 (2021).
Shi, X., Li, H. & Wang, H. Nutrient structure of the Taiwan Warm Current and estimation of vertical nutrient fluxes in upwelling areas in the East China Sea in summer. Journal of Ocean University of China. 13, 613–620 (2014).
Wang, W. et al. Intrusion pattern of the offshore Kuroshio branch current and its effects on nutrient contributions in the East China Sea. Journal of Geophysical Research: Oceans. 123, 2116–2128 (2018).
Liu, S. M. et al. Source versus recycling influences on the isotopic composition of nitrate and nitrite in the East China Sea. Journal of Geophysical Research: Oceans. 125, e2020JC016061 (2020).
Gu, X. et al. Dissolved nitrous oxide and hydroxylamine in the South Yellow Sea and the East China Sea during early spring: Distribution, production, and emissions. Frontiers in Marine Science. 8, 725713 (2021).
Zhu, L. et al. Estimate of dry deposition fluxes of nutrients over the East China Sea: The implication of aerosol ammonium to non-sea-salt sulfate ratio to nutrient deposition of coastal oceans. Atmospheric Environment. 69, 131–138 (2013).
Deng, X. et al. Carbonate chemistry variability in the southern Yellow Sea and East China Sea during spring of 2017 and summer of 2018. Science of The Total Environment. 779, 146376 (2021).
Ding, L., Ge, T. & Wang, X. Dissolved organic carbon dynamics in the East China Sea and the northwest Pacific Ocean. Ocean Science. 15, 1177–1190 (2019).
Wang, S. L. et al. Hypoxic effects on the radiocarbon in DIC of the ECS subsurface water. Journal of Geophysical Research: Oceans. 126, e2020JC016979 (2021).
Chou, W.-C. et al. The carbonate system in the East China Sea in winter. Marine Chemistry. 123, 44–55 (2011).
Sheu, D. D. et al. Riding over the Kuroshio from the South to the East China Sea: Mixing and transport of DIC. Geophysical research letters. 36, (2009).
Hung, J.-J., Lin, P.-L. & Liu, K.-K. Dissolved and particulate organic carbon in the southern East China Sea. Continental Shelf Research. 20, 545–569 (2000).
Meng, F. et al. Seasonal dynamics of dissolved organic carbon under complex circulation schemes on a large continental shelf: The Northern South China Sea. Journal of Geophysical Research: Oceans. 122, 9415–9428 (2017).
Hung, J.-J. et al. Distributions, stoichiometric patterns and cross-shelf exports of dissolved organic matter in the East China Sea. Deep Sea Research Part II: Topical Studies in Oceanography. 50, 1127–1145 (2003).
Kim, J. et al. Factors controlling the distributions of dissolved organic matter in the East China Sea during summer. Scientific reports. 10, 11854 (2020).
Liu, Q. et al. The satellite reversion of dissolved organic carbon (DOC) based on the analysis of the mixing behavior of DOC and colored dissolved organic matter: the East China Sea as an example. Acta Oceanologica Sinica. 32, 1–11 (2013).
Chen, C.-T. et al. Air–sea exchanges of CO 2 in the world’s coastal seas. Biogeosciences. 10, 6509–6544 (2013).
Zhu, Z. Y. et al. Bulk particulate organic carbon in the East China Sea: Tidal influence and bottom transport. Progress in Oceanography. 69, 37–60 (2006).
Wang, Y. et al. Seasonal variations in nutrients and biogenic particles in the upper and lower layers of East China Sea Shelf and their export to adjacent seas. Progress in Oceanography. 176, 102138 (2019).
Chen, Y.-lL. et al. New production in the East China Sea, comparison between well-mixed winter and stratified summer conditions. Continental Shelf Research. 21, 751–764 (2001).
Zhao, J. et al. Genetic variation of Ulva (Enteromorpha) prolifera (Ulvales, Chlorophyta)—the causative species of the green tides in the Yellow Sea, China. Journal of applied phycology. 23, 227–233 (2011).
Chen, Y.-L. L. & Chen, H.-Y. Seasonal dynamics of primary and new production in the northern South China Sea: The significance of river discharge and nutrient advection. Deep Sea Research Part I: Oceanographic Research Papers. 53, 971–986 (2006).
Acknowledgements
We gratefully acknowledge the very helpful comments from our two anonymous reviewers. We want to thank allthe staff of the R2R program, CCHDO, GLODAP and NASA Ocean Biology Distributed Active Archive Center(OB.DAAC) for enabling the access of the data. We also thank The Argo Data Management site, whose data arecollected and made freely available by the International Argo Program and the national programs that contributeto it. Meanwhile, acknowledgement for the data support from “National Earth System Science Data Center,National Science & Technology Infrastructure of China (http://www.geodata.cn)”. We thank Prof. Yuming Caifor providing the Chl a concentration of the Yellow Sea and East China Sea. We acknowledge Dr. Qiang Shi from the Dalhousie University for her help in improving the writing language of the revised manuscript. This research is funded by the National Key Research and Development Program of China (No. 2020YFA0608300 and No. 2024YFF0507000).
Author information
Authors and Affiliations
Contributions
B.S. conceived of the study. C.C.W. collected metadata, and compiled and analysed the dataset. C.C.W. and B.S. wrote the draft with inputs from J.S., X.K.H. and J.H.L. J.S. and X.K.H. provided in-situ Chl a concentration data of the CCS.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, C., Su, B., Sun, J. et al. A regional ocean database for the Coastal China Sea. Sci Data 12, 1550 (2025). https://doi.org/10.1038/s41597-025-05840-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-025-05840-w