Abstract
In this study we combined phenomics, proteomics, metabolomics and lipidomics analyses of historical and contemporary Bombyx mori cocoon shells to case-study the human-driven introduction and diversification of this species in Europe. Prompted by recent findings on the genomic variability that underlies the ancestry and cocoon shell colour of this species, we carried out optical and fluorescence imaging analysis of 148 cocoons shells to identify overt and covert phenotypic traits and employed LC-MS/MS analyses protocols for 80 cocoon shell samples to identify that the cocoon shell of this species contains on average 98 ± 13 (Mean ± SD) proteins, while we identified 141 metabolites and 981 lipids. We validated these generated datasets through multiple validation protocols, through a series of dimensionality reduction methods and clustering algorithms and through narratives from historical archives and manuscripts. Our multiomics datasets provide a valuable foundation for advancing further exploitations of silkworm cocoon shells in multiple scientific perspectives.
Similar content being viewed by others
Background & Summary
Recent studies1,2,3,4 on the evolutionary, phylogenetic diversity and speciation of the domesticated silkworm, Bombyx mori, have shown that this species has evolved from the Chinese wild silkworm, Bombyx mandarina, with the most recent common ancestor of the two species estimated to have existed about 4100 years ago1 in the territory north of the Qinling–Huaihe line in China1. Another analysis has indicated that the starting time of silkworm domestication was approximately 7500 years ago with the time of domestication termination estimated at 3984 years ago2. After its domestication, the silkworm underwent a population expansion within China during the Chinese Song Dynasty (960 CE–1279 CE)1, although evidently5,6, the silkworm was reared domestically before this time in regions outside East China. This latter point has been illustrated in a recent study of 1078 silkworm races and genetic stocks from China which documented the divergence of silkworm races across geographic regions and continents through genome sequencing3.
Several studies in the 20th century and a large cohort of silkworm rearing manuals7,8,9,10 accept that there are 4 different geographic clades of silkworm races: The Chinese races, the Japanese races, the Tropical races and the European silkworm races. The classification of the various silkworm races into four regional clades (Chinse, Japanese, European and Tropical)9 has its origins in the early 20th century10,11, it has become the main systematic classification tenet since the mid 20th century7,9, and continues to serve as a classification scheme even today3,12. This classification system serves its purpose well3 since in an impressive, large-scale genome sequencing study a clear segregation of the regional silkworm races was achieved through phylogenetic analysis of genome sequences. These authors showed that the European silkworm races that were used in their study, and were sourced from genetic stocks maintained within China, can be grouped into two branches: One branch is composed of races originating from republics of the former Soviet Union (but assigned as European) which are phylogenetically related to improved Japanese races and another tight and very early diverging branch which is composed of races from Central Europe and the Mediterranean Basin. This latter and early divergent branch of European silkworm races3 is quite coherent in its clustering with little phylogenetic similarity with the other extant Chinese silkworm races, an indication that a branch of Chinese silkworm races or a single Chinese race that does not exist in China anymore was the ancestor of all the European silkworm races much earlier than the genomic expansion of the silkworm races that took place during the time of the Song Dynasty in China1. This3, and a few other related studies, however, impressively detailed as they are3,4,12, did not sample any silkworm races from the Middle East so the actual early branching of the European silkworm races can still be considered an unresolved issue. This is rightly so since the contemporary West Asia silkworm races13 exhibit a large degree of admix while several historical and political events14 have probably resulted in their irrecoverable loss.
The results reported by Tong et al.3 served as one of the key motivations for generating the datasets presented in this study. Our aim was to provide the scientific community with tools to investigate the phenotypic diversity and historical origins of European silkworm races - a clade with obscure and unresolved ancestry9,10,15,16.
A further motivation was to establish an analytical framework for non-molecular phylogenetic analysis of both historical and contemporary biological specimens, particularly those for which genomic data cannot be retrieved, but other multiomics datasets can be generated17,18.
Yet another source of motivation was to provide meaningful support to museum collections and exhibitions of silkworm specimens by enriching them with analytical and historical context. The silkworm cocoon shell is a durable material that can endure for centuries under proper preservation and is often the only tangible remnant of silkworm specimens in museum collections. However, its historical significance is frequently underappreciated or poorly highlighted. The analytical framework presented in this study offers a paradigm for how multiomics approaches can advance the cultural and civic functions of museums in contemporary society.
In this study, we combined phenomic, proteomic, metabolomic, and lipidomic approaches to analyze historical silkworm cocoon specimens - alongside contemporary ones- where only the cocoon shell was preserved. The resulting datasets were processed using multiple clustering methods, and the outcomes were contextualized through historical texts and archival records to validate the analyses. Our analyses and validation protocols demonstrate that, even in the absence of genomic data, a multiomics strategy combined with robust clustering analysis can reveal hidden phenotypic traits and uncover previously unrecognized non-genomic phylogenetic relationships.
Methods
B. mori cocoon shell collection
A collection of 148 samples of cocoon shells from various, mostly European silkworm races, old silkworm races from various regions from or outside Europe and silkworm cocoon shell colour mutants was assembled from a diverse array of museums, private cocoon collections or genomic resources centres19 (Fig. 1a and details in Acknowledgements section). Use of cocoon shell samples was mandated by two facts: First, all the old cocoon shells, that we identified and collected, contained no remnants of the silkworm pupa in them and, secondly, national laws and legislations prohibited the export of genetic materials in other countries. Thereby, for the contemporary cocoon shell samples, the depositors were asked to send the cocoon shell samples upon removing any remnants of the silkworm pupae and larval exuviae from within the cocoon and handle the cocoon shells with sterile gloves to avoid any source of contamination. Despite the fact that the depositors were asked to handle cocoon shells with sterile gloves before shipment, contamination from human hands can not be ruled out in the case of old cocoon shell samples.
B. mori cocoon shell phenomics. (a) A worlds atlas showing by red circles the places of origin of the 148 cocoon shell samples that were used in this study. Many samples have an overlapping location and therefore are not shown (see19 for details). The inset image shows the places of origin of the European cocoon shell samples in more detail. (b) Adjusted absorbance measurements of all the cocoon shell samples. (c) Adjusted fluorescence spectra of all the cocoon shell samples upon excitation at 250 nm. Arrows indicate the peaks of fluorescence intensity of various cocoon shell samples. (d) Colour images of the 148 cocoon shell samples (upper row) and fluorescent grayscale images of the cocoon shells upon exposure to 365 nm UV light (lower row). Numbers indicate each sample’s ID19.
The collection comprised of 76 samples from contemporary cocoons produced between 2020–2023 while 72 samples were from old cocoons with the oldest dated in the early 19th century19. 50 of the 148 samples were phenotypically coloured while 88 were registered, upon receipt, as white19. Finally, 24 cocoon shell samples were of Asian origin, 2 samples were from Mexico (assigned as Asian for clustering purposes) and 122 samples were of European origin, all irrespective of where the cocoon shells were actually produced19. The depositors were asked to provide any further information about the cocoon shell samples, however, the information provided for the old cocoon shell samples were at certain instances limited19.
Phenomics analysis of B. mori cocoon shell
Biometric measurements of the cocoon shell samples were taken with a digital caliper for cocoon length, cocoon width and cocoon shell thickness for every cocoon in each sample20. To measure the absorbance spectra of the cocoon shells, a circular 7 mm piece of a cocoon shell was placed inside a well of a transparent microtiter plate (Corning). To measure the fluorescence spectra of the cocoon shells, the same circular 7 mm piece of the cocoon shell was transferred and placed inside a well of an opaque microtiter plate (Corning). Measurements were taken on a FlexStaion3 Multi-Mode Microplate Reader (Molecular Devices) in the fluorescence or absorbance spectra mode at every 5 nm from 200 nm to 990 nm. Absorbance and fluorescence scanning measurements were taken from the 148 samples of cocoon shells and upon correction for cocoon shell thickness using the equation: Aadjusted = Ameasured⋅ l0/l (mm), where l0 = 1 mm, the absorbance or fluorescence values were expressed as absorbance or fluorescence per 1 mm because there was a very wide variability in cocoon shell thickness.
Imaging of B. mori cocoon shells
Images of cocoons were taken with a Canon EOS 30D camera from a distance of 20 cm within a white illumination box. The camera settings were: AEB 0, WB SHIFT/BKT: 0, 0/ ± 0, Colour temp.: 4000 K, Colour space: Adobe RGB. A colour coded ruler was place next to the cocoon shell samples for size indication. Grayscale fluorescent imaging was taken by placing the cocoon shell in a UV illumination chamber (Axygen® Gel Documentation System, Corning) and collecting images upon exposure at 365 nm after a 2 sec exposure. Fluorescent colour imaging was taken with the above-mentioned camera in A-DEP mode from a distance of 20 cm while the cocoons were placed on a 365 nm UV-transilluminator. The fluorescent colour images were processed in Adobe Photoshop 20.0.3 (Adobe) to turn the background colour to black (RGB values set to 0, 0, 0) and further cropped. All the raw images of the cocoon shells were only cropped and for colour approximation of the old cocoon shell samples (See Validation section), only the cocoon shells RGB and HSB values were adjusted to show the estimated original appearance of the cocoon shells upon calculation of their actual original colour. The RGB and HSB values of the images were measured using Fiji21 with measurements taken from a circular area of 5 × 5 mm on one of the cocoons of each sample.
Original image approximation for the old cocoon shell samples was carried out using multi-linear regression analysis of the contemporary cocoon shell samples and was applied to the old cocoon shell sample (See Validation section). The equations used were:
Chemicals
LC-MS grade water, acetonitrile (ACN), methanol (MeOH), and isopropanol (IPA) were obtained from Th. Geyer. High-purity methyl tert-butyl ether (MTBE), ammonium formate, formic acid, ammonium acetate, and acetic acid were purchased from Merck. Stable isotope labelled internal standards for lipidomics (EquiSPLASH; Avanti Polar Lipids) and metabolomics (MSK-A2-1.2; Cambridge Isotope Laboratories) were used at final concentrations of 0.5% and 1.0% (v/v), respectively. Rutin (quercetin-3-O-rutinoside) quercetin and riboflavin were purchased from Cayman Chemical. Analytical grade oleic acid was purchased from Sigma-Aldrich. 1-amino-2-naphthol-4-sulfonic acid was purchased from Merck. Sodium bisulfite (NaHSO3), sodium sulfite (Na2SO3), ammonium molybdate and perchloric acid were purchased from Sigma-Aldrich. Sulfuric acid and orthophosphoric acid were purchased from Thermo Fisher Scientific.
Cocoon shell sample preparation for metabolomics and lipidomics analysis
A set of 8022 of the 148 cocoon shell samples were selected for metabolomics and lipidomics analysis. A pilot study was carried out with 4 additional samples to validate every step of the sample analysis and assess its feasibility. The results of the pilot study are not included in the analysis of the 80 samples. A careful selection of the 80 cocoon shell samples was carried out aiming to avoid as much as possible ambiguously-labelled B. mori mutants22, cocoon shell samples of uncertain origin, research purpose-generated mutants as well as samples with known parental-sibling relationships (i.e. samples 141–14722) that would skew the analysis results. Purposefully, samples of the same name or similar geographic location were used in the analysis to assess the effect of the place of origin and sample freshness on the obtained results.
For biphasic extraction of lipids and polar metabolites, cocoons were cut into small pieces and placed in 5 mL centrifuge tubes. Three (3) mL of MeOH (containing internal standards) were initially added to the cocoon samples. After vortexing and ultrasonic bath extraction for 10 min, 1.8 mL of supernatant were collected and transferred to clean 15 mL centrifuge tubes. Subsequently, 4.5 mL of MTBE were added and the monophasic mixture was vortexed for 60 s and incubated at −20 °C for 20 min. For phase separation, 1.2 mL water were added, followed by another vortexing and incubation step (see previous conditions). The biphasic solvent system was then centrifuged for 15 min at 5000 rpm and 4 °C with a 5810 R centrifuge (Eppendorf). For lipidomics analysis, 4 mL of the upper organic phase were transferred, dried under a stream of nitrogen, and reconstituted in 100 µL IPA:MeOH (50:50, v/v). For metabolomics analysis, 2.4 mL of the lower aqueous phase were transferred, dried under a stream of nitrogen, and reconstituted in 75 µL 80% MeOH (v/v). The final samples were vortexed for 10 min, centrifuged (see previous conditions) and the supernatants were transferred to analytical glass vials for LC-MS/MS analysis.
LC-MS/MS Instrumentation
LC-MS/MS analysis was performed on a Vanquish UHPLC system coupled to an Orbitrap Exploris 240 high-resolution mass spectrometer (Thermo Fisher Scientific) in positive and negative HESI (Heated ElectroSpray Ionization) mode.
Untargeted metabolomics analysis
Chromatographic separation was carried out on an Atlantis Premier BEH Z-HILIC column (Waters; 2.1 mm × 100 mm, 1.7 µm) at a flow rate of 0.25 mL/min. The mobile phase consisted of water:acetonitrile (9:1, v/v; mobile phase A) and acetonitrile:water (9:1, v/v; mobile phase B), which were modified with a total buffer concentration of 10 mM ammonium acetate (negative mode) and 10 mM ammonium formate (positive mode), respectively. The aqueous portion of each mobile phase was pH-adjusted (negative mode: pH 9.0) via addition of ammonium hydroxide and positive mode: pH 3.0 via addition of formic acid. The following gradient (20 min total run time including re-equilibration) was applied (time [min]/%B): 0/95, 2/95, 14.5/60, 16/60, 16.5/95, 20/95. Column temperature was maintained at 40 °C, the autosampler was set to 4 °C and sample injection volume was 5 µL.
Analytes were recorded via a full scan with a mass resolving power of 120 K over a mass range from 60–900 m/z (scan time: 100 ms, RF lens: 70%). To obtain MS/MS fragment spectra, data-dependant acquisition was carried out (resolving power: 15,000; scan time: 22 ms; stepped collision energies [%]: 30/50/70; cycle time: 900 ms). Ion source parameters were set to the following values: spray voltage: 4100 V (positive mode)/ −3500 V (negative mode), sheath gas: 30 psi, arbitrary units, auxiliary gas: 5 psi, arbitrary units, sweep gas: 0 psi, arbitrary units, ion transfer tube temperature: 350 °C, vaporizer temperature: 300 °C.
Untargeted lipidomics analysis
Chromatographic separation was carried out on an ACQUITY Premier CSH C18 column (Waters; 2.1 mm × 100 mm, 1.7 µm) at a flow rate of 0.3 mL/min. The mobile phase consisted of water:acetonitrile (40:60, v/v; mobile phase A) and isopropanol:acetonitrile (9:1, v/v; mobile phase B), which were modified with a total buffer concentration of 10 mM ammonium acetate + 0.1% acetic acid (negative mode) and 10 mM ammonium formate + 0.1% formic acid (positive mode), respectively. The following gradient (23 min total run time including re-equilibration) was applied (min/%B): 0/15, 2.5/30, 3.2/48, 15/82, 17.5/99, 19.5/99, 20/15, 23/15. Column temperature was maintained at 65 °C, the autosampler was set to 4 °C and sample injection volume was 5 µL.
Analytes were recorded via a full scan with a mass resolving power of 120 K over a mass range from 200–1700 m/z (scan time: 100 ms, RF lens: 70%). To obtain MS/MS fragment spectra, data-dependant acquisition was carried out (resolving power: 15,000; scan time: 54 ms; stepped collision energies [%]: 25/35/50; cycle time: 600 ms). Ion source parameters were set to the following values: spray voltage: 3250 V/-3000 V, sheath gas: 45 psi, auxiliary gas: 15 psi, sweep gas: 0 psi, ion transfer tube temperature: 300 °C, vaporizer temperature: 275 °C.
For both the metabolomics (see previous section) and the lipidomics analyses, all experimental samples were measured in a randomized manner. In both analyses, pooled quality control (QC) samples were prepared by mixing equal aliquots from each processed sample. Multiple QCs were injected at the beginning of the analysis to equilibrate the analytical system. A QC sample was analysed after every 5th experimental sample to monitor instrument performance throughout the sequence23.
For determination of background signals and subsequent background subtraction, an additional processed blank sample was recorded. Data was processed using MS-DIAL 4.9.22121824 and raw peak intensity data was normalized via total ion count of all detected analytes25. Feature identification was based on the MS-DIAL LipidBlast V68 library26 (lipidomics) and Level 1 feature identification was employed for the in-house library for metabolomics (EMBL-MCF 2.0) using accurate mass, isotope pattern, MS/MS fragmentation, retention time information and a minimum matching score of 80% in MS-DIAL software.
Cocoon shell sample processing for proteomics analysis
For Sp3-mediated protein digestion27, pieces of 20 mg of cocoon shells of each sample were further cut into smaller pieces and immersed into 400 μL lysis buffer (4% sodium dodecyl sulfate, 100 mM dithiothreitol and 100 mM triethylammonium bicarbonate (pH = 8.5)). The mixtures were subjected to two alternating rounds of heating at 99 °C for 5 mn. and ultrasonic bath for 10 min. Finally, the samples were centrifuged at 15000xg for 15 min and the supernatant of each sample was processed according to the Single-Pot Solid-Phase-enhanced Sample Preparation (Sp3) protocol27, without acidification and including protein and cysteine alkylation steps in 100 mM iodoacetamide. Proteins in the samples were allowed to bind to 20 μg of beads (1:1 mixture of hydrophilic and hydrophobic SeraMag carboxylate-modified beads, (GE Life Sciences) in 50% ethanol for 15 min. Using a magnetic rack, beads were washed twice with 80% ethanol and once with 100% acetonitrile. Protease cleavage of the solubilized proteins was carried out by continuous shaking at 1200 rpm at 37 °C for 24 hrs using 0.5 μg Trypsin Platinum, Mass Spectrometry Grade (Promega) in a 100 mM triethylammonium bicarbonate buffer (pH = 8.5). The next day, peptides were further purified by Sp3 peptide clean-up and evaporated to dryness in a vacuum centrifuge. The dried samples were solubilized in mobile phase A (2% acetonitrile,0.1% formic acid in LC-MS water), sonicated for 5 min and the peptide concentration was determined by measuring the absorbance at 280 nm using a nanodrop.
Proteomic LC-MS/MS analysis
Samples were run on a liquid chromatography tandem mass spectrometry (LC-MS/MS) setup consisting of a Dionex Ultimate 3000RSLC online with a Thermo Q Exactive HF-X Orbitrap mass spectrometer. Peptidic samples were directly injected and separated on a 25 cm-long analytical C18 column (PepSep, 1.9μm3 beads, 75 µm ID) using a 1-hour long run, starting with a gradient of 7% Buffer B (0.1% formic acid in 80% acetonitrile) to 35% for 40 min and followed by an increase to 45% in 5 min and a second increase to 99% in 0.5 min and then kept constant for equilibration for 14.5 min. A full MS was acquired in profile mode using a Q Exactive HF-X Hybrid Quadropole-Orbitrap mass spectrometer, operating in the scan range of 375–1400 m/z using 120 K resolving power with an AGC of 3 × 106 and max IT of 60 ms followed by data independent analysis using 8 Th windows (39 loop counts) with 15 K resolving power with an AGC of 3 × 105 and max IT of 22 ms and a normalized collision energy (NCE) of 26. Each biological sample was analysed in two technical replicas on the system.
Proteomic data analysis
Orbitrap raw data was analysed in DIA-NN 1.9.2 (Data-Independent Acquisition by Neural Networks)28 through searching against three B. mori proteome datasets retrieved from Uniprot or NCBI or Kaikobase in the library free mode of the software, allowing up to two tryptic missed cleavages. A spectral library was created from the DIA runs and used to reanalyse them. DIA-NN default settings have been used with oxidation of methionine residues and acetylation of the protein N-termini set as variable modifications and carbamidomethylation of cysteine residues as fixed modification29. N-terminal methionine excision was also enabled. The match between runs (MBR) feature was used for all analyses and the output (precursor) was filtered at 1% FDR and finally the protein inference was performed on the level of genes using only proteotypic peptides. The generated results were processed statistically and visualized in the Perseus software (1.6.15.0)30.
Monoclonal antibody generation for proteomic analysis validation
To validate the proteomic analysis results, a mouse monoclonal antibody was generated against a peptide (N– NENLDIDRTHDNYRC–C) corresponding to amino acids 65–78 of B. mori imaginal disk growth factor protein (GenBank acc. no.: BAF73623). The B. mori imaginal disk growth factor is a 434 amino acid protein that contains a 16-amino acid secretory signal peptide at its N-terminal and the mature secreted protein has a MW of 46.50 kDa. With a cysteine attached to its N-terminal, the 15 amino acid peptide was conjugated to keyhole limpet hemocyanin (KLH) and the conjugate was used as antigen. This antigen was produced and purified by GenScript. Monoclonal antibodies against the same peptide were produced according to a modified method31,32. Five BALB/c mice of 5 weeks of age were immunised intraperitoneally (i.p.) with 25 μg of the KLH-conjugated peptide. All immunisation and animal handling were in accordance with animal care guidelines as specified in EU Directive 2010/63/EU. After 5 cycles of immunisation, mice were euthanised and spleenocytes were collected and fused with the P3X63Ag8.653 cell line (ATCC® CRL1580™) according to the fusion protocol31. Positive clones and antibody specificity were determined through immunoblotting and immunosorbent assays and, among the several positive clones, one was further propagated and used.
Western blots
A sample of 25 μL of the homogenate that was used for proteomics analysis was mixed with a 6x protein samples loading buffer (Thermo Fisher Scientific) and resolved in a 10% acrylamide/bis-acrylamide SDS-PAGE gel. Gels were wet transferred onto a PVDF membrane (Millipore) in transfer buffer (25 mmol tris, 192 mmol glycine, 20% (v/v) methanol) at 100 V constant at 4 °C for 90 min. PVDF membranes were blocked by incubation with 5% BSA in TBST (25 mmol tris-HCl, (pH = 7.5), 150 mmol NaCl, 0.1% tween 20) for 1 h at room temperature32. Membranes were then incubated for 1 h at room temperature with the mouse anti-imaginal disk growth factor monoclonal antibody at 1:1000 dilution of the cloned cell line culture supernatant in TBST with 5% BSA. Membranes were then washed with TBST (3 × 5 min) and incubated with an HRP-conjugated anti-mouse antibody (1:1000, Jackson Laboratories) for 1 h at room temperature. Immunoreactive bands were detected using the Luminata Crescendo HRP substrate (Millipore) in an Alpha Innotech FluorChem 8800 imaging system.
Determination of total flavonoids in cocoon shells
Determination of total flavonoids in cocoon shells followed the protocol of Lu et al.12. 10 mg of a cocoon shell from all the 148 cocoon shell samples was cut in smaller pieces, immersed in a 200 μL solution of 40% ethanol so that the ratio of solid to liquid was 1:20, and stored overnight at −80 °C. Then, the solution was sonicated for 20 min and 100 μL were carefully removed and absorbance measurements at 355 nm were taken on a FlexStaion3 Multi-Mode Microplate Reader (Molecular Devices). Standard curves were constructed for rutin, quercetin and riboflavin which had absorbance maxima at 355, 355 and 370 nm, respectively, while riboflavin showed an additional absorbance peak at 455 nm. Rutin had the best dynamic range of the three compounds over a range of 1000-fold dilution and so total flavonoids content of cocoon shell samples was expressed as rutin equivalents since rutin was used as the standard for constructing the calibration curve.
Determination of free amino acids in cocoon shells
To validate the identification of free amino acids in the cocoon shells as identified by the metabolomics analysis, the same protocol for sample preparation to that applied for metabolomics and lipidomics analysis was employed. The only differences with the above-mentioned protocol were: 1) the extraction volume was scaled down by 10 times, 2) The analysis of free amino acids was carried out in a piece of 10 mg of cocoon shell and not in whole cocoon shells and 3) for validation purposes the analysis was carried out in 20 cocoon shell samples (10 contemporary and 10 old cocoon shell samples). A 10 μL of the lower aqueous phase of each cocoon shell extract was mixed with 90 μL acetonitrile and a volume of 20 μL of this mixture was analysed using liquid chromatography coupled to a triple quadrupole mass spectrometer (Sciex Qtrap 5500 + (AB Sciex)33. Chromatographic separation was performed using an ACQUITY BEH amide column (Waters; 150 mm × 2.1 mm, 1.7 μm), and the mobile phase consisted of (A) acetonitrile, formic acid 0.15% and (B) 5 mM ammonium formate, formic acid 0.15%. Selected reaction monitoring (SRM) with electrospray ionization (ESI) in positive mode was applied34.
Determination of various classes of lipids in cocoon shells
While the aqueous layer was used for determination of free amino acids as described above, the upper organic layer was used for the quantitative determination of lipids in the cocoon cells. We employed two different assays for the quantification of lipids in cocoon shells. The first assay was a modification of the Bartlett’s35 assay for the determination of phospholipids. The assay protocol was as follows: 100 μL of samples in MTBE were left to evaporate and then incubated with 400 μL 70% perchloric acid for 30 min at 180 °C until a clear solution was formed. Then 1.2 mL HPLC-grade ddH2O was added followed by addition of 0.2 mL 5% ammonium molybdate solution in HPLC-grade ddH2O. Upon mixing and storing at room temperature for 5 min, 50 μL of freshly prepared Fiske–Subbarow Reducing Reagent was added This reagent is composed of 0.2 g 1-amino-2-naphthol-4-sulfonic acid, 1.2 g sodium bisulfite (NaHSO3) and 1.2 g sodium sulfite (Na2SO3)36 dissolved in 100 mL of HPLC-grade ddH2O. After incubation for 10 min at room temperature, absorbance was read at 830 nm on a 96-well microtiter plate (Corning) using the FlexStaion3 Multi-Mode Microplate Reader (Molecular Devices). Potassium dihydrogen phosphate (KH2PO4)37,38 was used to generate the standard curve and the results are expressed as μg P/cocoon shell weight.
For the determination of total amount of lipids we used the sulfo-phospho-vanillin assay39 as follows: 20 μL of the upper organic layer of the extracted samples were evaporated at room temperature and then the lipid film that was formed was dissolved in 200 μL concentrated sulfuric acid and heated to 100 °C for 10 min. Following cooling at room temperature for 15 min, 0.5 mL of vanillin-orthophosphoric acid reagent was added. The phosphovanilin reagent was prepared by dissolving 0.2 g of vanillin (AppliChem) into 20 mL of hot ddH2O (70 °C) and further diluted to 100 mL with 85% orthophosphoric acid while stirring the solution. The samples were incubated at 37 °C for 15 min and then stored for 45 min in the dark before measuring absorbance at 530 nm on a 96-well microtiter plate (Corning) using the FlexStaion3 Multi-Mode Microplate Reader (Molecular Devices). Oleic acid (Sigma-Aldrich) was used as a calibration standard and had an EC50 of 8.6 μg in this assay setup.
Bioinformatics
The analysis platform of Metaboanalyst 6.0 (https://www.metaboanalyst.ca/) was employed to perform multivariate data analysis of the acquired lipidomics and metabolomics data. The signal intensity data of the analysed samples and the pooled quality control (QC) samples was used in both cases for any subsequent analysis without any further normalization. Among the several chemometric techniques on Metaboanalyst 6.0, the partial least squares discriminant analysis (PLS-DA) provided the most discriminatory results40 in identifying the metabolomic differences among samples, particularly in distinguishing different metabolites or lipids and 5% FDR was the criteria used to define clusters. For GO term enrichment analysis of the identified proteins in cocoon shell samples, ShinyGO 0.8241 (https://bioinformatics.sdstate.edu/go/) was used. An FDR value of 5% was set and the q-values are shown in Log10 scale for clarity.
Computational analytical methods
Clustered heatmaps of the proteomics, metabolomics, and lipidomics datasets were created using the NG-CHM Builder42 with the Euclidean distance matrix and Ward’s agglomeration43. The best visual resolution was achieved with seven clusters of samples and nine clusters of either proteins, metabolites, or lipids in the Euclidean distance metric and Ward’s algorithm. The spreadsheets44,45,46 containing the identified metabolites and lipids, from both the negative and positive mode runs, were merged for this analysis. If a lipid or metabolite was identified in both modes, the one with the weaker detection signal was discarded. Next, for each analysed sample, the total signal intensity was summed. Then, for each metabolite or lipid, the ratio of its intensity to the total intensity of the sample was calculated. This ratio was subsequently normalized to 100 mg of cocoon shell, as each cocoon had a different shell weight20. For Decision Tree analyses47, we used the CART decision tree classifier48.
A variety of dimensionality reduction methods49,50, such as the t-distributed Stochastic Neighbor Embedding (t-SNE)51, the Uniform Manifold Approximation and Projection (UMAP)52, the Principal Component Analysis (PCA), the MultiDimensional Scaling (MDS)53, the Isometric Mapping (ISOMAP)54 and the Locally Linear Embedding (LLE)55,56 were used57 to reduce the dimensionality of the proteomics, metabolomics and lipidomics datasets and provide a graphical overview of the datasets either in a two-dimensional49 or three-dimensional50 space. The three-dimensional representations of the proteomics, metabolomics and lipidomics datasets can be viewed in50, however, not all methods provided clear results by resolving the samples in a two dimensional49 or three dimensional space50. Among the methods used49,50, Locally Linear Embedding (LLE) consistently provided the clearest results. Locally Linear Embedding (LLE)55,56 is a nonlinear dimensionality reduction technique that helps visualize high-dimensional data in lower dimensions. When applied to two-dimensional plots, LLE provides a compressed yet informative representation of the original data, preserving local or global structures55,56. Locally Linear Embedding captures local neighbourhood relationships, making it useful for discovering intrinsic manifold structures. When this method is used to generate three-dimensional plots50, an additional degree of freedom is introduced, allowing for a more detailed representation of the manifold structure, potentially reducing distortions that occur in two-dimensional projections. This is particularly useful when the intrinsic dimensionality of the data is closer to three, as it enables better separation of clusters or curved structures that might overlap in a two-dimensional space.
Statistical analysis
All data that is presented in our study were analysed using SPSS v28 (IBM Corp.) or GraphPad Prism v.10. Multiple t-test analyses and non-linear regression analyses were carried out in GraphPad Prism v.10 to generate the false discovery rate adjusted p-value of 0.01. Non-linear regression analyses and determination of the amounts of lipids in cocoon shells was also carried out on GraphPad Prism v.10.
Data Records
The raw files and annotated data of the metabolomics and lipidomics analyses of the 80 cocoon shell samples58 are available at the NIH Common Fund’s National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org59 where they have been assigned Study ID ST003842 and ST003843, respectively. The data can be accessed directly via its Project https://doi.org/10.21228/M88V68.
The mass spectrometry proteomics raw data have been deposited on the ProteomeXchange Consortium via the PRIDE (https://www.ebi.ac.uk/pride/)60 partner repository with the dataset identifier PXD06235161.
The B. mori cocoon shell collection descriptor of the 148 cocoon shell samples19 in .xlsx format is available on Figshare (https://doi.org/10.6084/m9.figshare.29974921).
The lists of B. mori cocoon shell samples selected (80 samples) and excluded (68 samples) for proteomics, metabolomics and lipidomicds analyses and their selection justification22 in .xlsx format are available on Figshare (https://doi.org/10.6084/m9.figshare.29975101).
The phenomics and biometric data of the 148 cocoon shells samples20 in .xlsx format is available on Figshare (https://doi.org/10.6084/m9.figshare.29975173).
The adjusted absorbance measurements of the 148 cocoon shells samples62 in .xlsx format are available on Figshare (https://doi.org/10.6084/m9.figshare.29975215).
The fluorescence intensity measurements of 47 cocoon shell samples63 in .xlsx format are available on Figshare (https://doi.org/10.6084/m9.figshare.29975242).
The phenomics data of the148 cocoon shells samples (RGB and HSB values)64 in .xlsx format is available on Figshare (https://doi.org/10.6084/m9.figshare.29975275).
The adjusted phenomics data of the148 cocoon shells samples (original and adjusted RGB and HSB values) based on the non-linear regression equations65 in .xlsx format is available on Figshare (https://doi.org/10.6084/m9.figshare.29975305).
The clustering data of the148 cocoon shells samples based on phenomics analysis66 in .xlsx forma is available on Figshare (https://doi.org/10.6084/m9.figshare.29975329).
The proteomics results for 81 B. mori cocoon shell samples45 in .xlsx format are available on Figshare (https://doi.org/10.6084/m9.figshare.29975422).
The metabolomics results for 80 B. mori cocoon shell samples44 in .xlsx format, along with 4 additional files in .txt format containing the raw data results (including QC values) and the Total Ion Current (TIC)-normalized results for both the positive and negative modes, are available on Figshare (https://doi.org/10.6084/m9.figshare.29975479).
The raw data of the free amino acids validation analysis run on Sciex Qtrap 5500+ in .wiff and .wiff.scan format33 is available on Figshare (https://doi.org/10.6084/m9.figshare.29975932).
The lipidomics results for 80 B. mori cocoon shell samples46 in .xlsx format, along with 4 additional files in .txt format containing the raw data results (including QC values) and the Total Ion Current (TIC)-normalized results for both the positive and negative modes, are available on Figshare (https://doi.org/10.6084/m9.figshare.29975560).
The decision trees analysis results for the proteomics, metabolomics and lipidomics datasets47 in .tiff format are available on Figshare (https://doi.org/10.6084/m9.figshare.29975653).
The PLS-DA analysis results for the metabolomics and lipidomics datasets40 in .png format are available on Figshare (https://doi.org/10.6084/m9.figshare.29976385).
The custom Google Colab executable codes57 in .py format are available on Figshare (https://doi.org/10.6084/m9.figshare.28710992).
The 2D dimensionality reduction plots for the proteomics, metabolomics and lipidomics datasets49 in .png format are available on Figshare (https://doi.org/10.6084/m9.figshare.29976331).
The 3D interactive dimensionality reduction plots for the proteomics, metabolomics and lipidomics datasets50 in .html format are available on Figshare (https://doi.org/10.6084/m9.figshare.27985811).
The hierarchical clustering analysis results for the proteomics, metabolomics and lipidomics datasets67 in .png format are available on Figshare (https://doi.org/10.6084/m9.figshare.29976298).
The k-means clustering results of the proteomics, metabolomics and lipidomics datasets68 in .xlsx format are available on Figshare (https://doi.org/10.6084/m9.figshare.29975617).
The Silhouette values of the k-means analysis results for the proteomics, metabolomics and lipidomics datasets69 in .png format are available on Figshare (https://doi.org/10.6084/m9.figshare.29976445).
Technical Validation
Validation of cocoon shell phenomics and colour approximation
The B. mori cocoon shells exhibit variations in absorbance between 280–560 nm (Fig. 1b and62). These inconsistencies observed in adjusted absorbance scanning measurements (Fig. 1b) were related to cocoon age62 and this prompted examination of their fluorescence spectra (Fig. 1c), since it was shown that B. mori cocoon shells have intrinsic fluorescence12. In addition to fluorescence intensity peaks at 550 nm (Fig. 1c)70, some samples exhibited peaks at 480–490 nm, others at 450–460 nm and others at 440 nm (Fig. 1c). To corroborate the results in Fig. 1c, we followed the protocol of Lu et al.12 and recorded greyscale images of the cocoon shells upon UV irradiation at 365 nm. Forty-six of the 148 samples displayed fluorescence (Fig. 1d)12,70. Fluorescence intensity was quantified63 in Fiji21 to corroborate the image data in Fig. 1d. Because visible pigments were not detectable in old cocoon shell samples (Fig. 1d), colour approximation methods were applied (see Methods).
To approximately reconstitute the original colour of the old cocoon shell samples and enable downstream validation, we applied multi-linear regression analysis using the RGB and HSB values of contemporary cocoon shells64 and transferred the calculated values to the old samples as follows: First, the Green (G) channel values were plotted against Brightness for contemporary cocoons, and five groups were resolved, with white cocoon shells forming a linear relationship (Fig. 2a and64. Next, Blue (B) channel values of the contemporary cocoons plotted against Saturation showed a negative correlation with five groups forming, while white cocoons had negligible saturation values (<0.1; Fig. 2b). In addition, Hue versus Saturation values of the contemporary cocoons also produced five groups, with six cocoon shell samples having Hue > 150° and the rest between 40–60° (Fig. 2c). Old cocoon shell samples appeared faint white to off-white with RGB values (214.1 ± 12.3, 212.1 ± 13, 199 ± 19.6 (Mean ± SD) and HSB values (56 ± 7.7, 0.07 ± 0.05, 0.84 ± 0.05 (Mean ± SD) (N = 72)64. When Green (G) channel values of old cocoons were plotted against Brightness (Fig. 2d), the old cocoon shells showed a similar trend to contemporary samples (compare Fig. 2a,d). Plots of Blue (B) channel against Saturation of old cocoons (Fig. 2e) showed a modest negative correlation, reflecting the generally low Blue channel values in old cocoons. Hue versus Saturation values of old cocoons (Fig. 2f) indicated that all old samples had Hue values between 40–60°.
B. mori cocoon shell colour phenomics. (a) Correlation in contemporary cocoon shells between Green channel values and Brightness values on the RGB and HSB scales64. For colour coding of the samples in this and all subsequent panels of Fig. 2, see65. (b) Correlation in the contemporary cocoon shells between Blue channel values and Saturation values. (c) Correlation in the contemporary cocoon shells between Hue and Saturation values on the HSB scale. The Hue values are expressed as point in the colour wheel. (d) Correlation in the old cocoon shells between Green channel values and Brightness values64 (e) Correlation in the old cocoon shells between Blue channel values and Saturation values. (f) Correlation in the old cocoon shells between Hue and Saturation values om the HSB scales. The Hue values are expressed as point in the colour wheel. (g) Correlation in the old cocoon shells between Green channel values and Brightness values after adjusting the original colour of the samples using the equations shown in Methods section. (h) Correlation in the old cocoon shells between Blue channel values and Saturation values after adjusting the original colour of the samples using the equations shown in Methods section. (i) Correlation in the old cocoon shells between Hue and Saturation values om the HSB scales after adjusting the original colour of the samples using the equations shown in Methods section. (j) Reconstruction of the original colour of the old cocoon shell samples in Photoshop v. 20.0.3 (upper row) and fluorescent colour images of the cocoon shells upon exposure to 365 nm UV light (lower row). Numbers indicate each sample’s ID19. * Sample No. 142 had the lowest fluorescence intensity values of all the samples shown in Fig. 1d appearing extremely dark, with a Hue value of 220° and the lowest Blue channel value of all the samples in (j) The colour images of Fig. 1d were used to reconstruct their original colour in Photoshop v. 20.0.3. The original and adjusted RGB and HSB values are shown in65. (k) Clustering of all the cocoon shell samples to 8 distinct clusters based on their colour appearance upon UV light exposure as shown in (j). The samples were classified according to the presence of green pigments on the cocoon shell, their original colour (White/Coloured) and the presence of fluorescence as recorded in Fig. 1d. Numbers indicate each sample’s ID19. Numbers in red indicate the old cocoon shell samples and numbers in black the contemporary cocoon shell samples.
Using the set of non-linear regression equations, original RGB and HSB values were calculated for the old cocoons65. We, then, plotted the calculated Green (G) channel values against the Brightness values (Fig. 2g), the calculated Blue (B) channel values against the Saturation values (Fig. 2h) and the Hue values against the calculated Saturation values (Fig. 2i). These calculated values were used to generate the approximate colour of the old cocoon shells (Fig. 2j). High-resolution colour images of each cocoon shell under UV irradiation at 365 nm were also obtained (Fig. 2j). The composite images show variable degrees of green pigmentation across several samples, and in many cases, old cocoons had strong green pigmentation (Fig. 2j).
Upon colour approximation of the old cocoons, all 148 cocoon shell samples were further grouped into clusters based on three parameters: (i) presence or absence of green pigments (Fig. 2j), (ii) presence or absence of fluorescence (Fig. 1d), and (iii) presence or absence of white colour, as determined by the calculated RGB and HSB values (Fig. 2a–i). Eight clusters of cocoon shells were resolved (Fig. 2k). Cluster 4 (white and fluorescent cocoons) contained no old cocoon shells, while Cluster 7 (no green pigments, non-white, non-fluorescent) contained most of the old cocoon shells19,66. Most contemporary cocoon shell samples were placed in Cluster 3 (Fig. 2k and19,66). Putative genotypes for cocoon colour were assigned to clusters based on references from Daimon et al.70 and Lu et al.12 (Fig. 2k and66). The calculated colour values (Fig. 2j) and cluster assignments (Fig. 2k) were subsequently used in downstream clustering validations of the proteomic, metabolomic, and lipidomic datasets, and were also compared with archival references for consistency.
Proteomic data validation
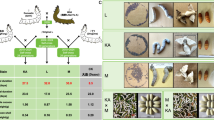
To validate the LC-MS/MS proteomics pipeline for the 81 cocoon shell samples analysed, results were first filtered (Fig. 3a) to remove erroneous annotations from the three B. mori genome databases used. A total of 233 proteins were identified. The majority (57 proteins, each present in < 5% of samples) were uniquely detected in individual samples45. Twenty-three proteins were consistently detected in all samples (Fig. 3a), and 39 proteins were identified in ≥ 95% of samples representing a conserved core set consistently detected in B. mori cocoon shells45. This identified protein set is consistent with previous proteomic studies of B. mori cocoon shells71,72,73,74,75,76 and includes all the well-characterized proteins that are structural components of the silk thread as well as several enzymes and immunity-related proteins45,71,72,73,74,75,76.
B. mori cocoon shell proteomics. (a) Heat map of signal intensity fractions of all the 233 identified proteins in all the 81 samples that were analysed. (b) GO term analysis of the enriched proteins in all the cocoon shell samples that were analysed. (c) Representative Western blot showing the single immunoreactive band of imaginal disk growth factor protein migrating at around 46 kDa in extracts from 6 cocoon shell samples (left side of the image) in which this protein was identified by proteomics analysis. In the right side of the image and next to the protein ladder, extracts from 3 cocoon shell samples were loaded in which this protein was not identified by proteomics analysis. The sample description indicates the age of the sample (C = Contemporary; O = Old), its sample ID19 and sample’s name. On the right side of the image, the migration of protein standards included in the protein ladder is shown.
Most of the large subset of immune response-related proteins were found in all the cocoon samples (Fig. 3a), while only 3 proteins composed the majority of the normalized protein abundance values (Fig. 3a). These 3 proteins are Sericin 1, Sericin 3 and Osiris 9A73.
The number of proteins identified per sample was also examined. Contemporary cocoon shells contained 102 ± 12 proteins (Mean ± SD), while old cocoon shells contained 96 ± 13 proteins (Mean ± SD). Across all samples, an average of 98 ± 13 proteins was identified per cocoon shell (Fig. 3a). Comparison with a set of 194 protein-encoding genes previously associated with cocoon yield77, showed that only one of these, general odorant-binding protein 70, was identified, and only in a single sample (Fig. 3a and45).
To further assess the dataset, GO term enrichment analysis of the identified proteins was carried out (Fig. 3b). The analysis revealed that many proteins belonged to immunity-related categories, including enzymes, small peptides, hydrolases, and protease inhibitors (Fig. 3b)74,75.
As an additional validation step, a monoclonal antibody was generated against a peptide fragment of the imaginal disk growth factor protein of B. mori, identified in 20 of the 81 cocoon shell samples45. This protein contains an N-terminal secretory signal peptide and has been previously characterized in B. mori78. Western blot analysis confirmed immunoreactive bands only in all 6 extracts of cocoon shells samples where the protein had been identified by proteomics analysis (Fig. 3c), and no signal was detected in samples where the protein was absent from the proteomics dataset45.
Metabolomic data validation
Metabolomic analysis of the same as above 80 cocoons shell samples identified 141 distinct metabolites as present in B. mori cocoon shells (Fig. 4a and44). The identified metabolites were classified using RefMet79. A Circos plot80 of metabolite super-classes (Fig. 4b) showed the predominant presence of free amino acids, along with additional metabolite classes, such as alkaloids44, detected across samples irrespective of cocoon age, origin, or colour (Fig. 4b). For example, urocanic acid, a chemical compound known to protect from UV-B irradiation81, was detected mainly in old Asian samples and much less in certain old Italian and French samples44.
B. mori cocoon shell metabolomics. (a) Heat map of signal intensity fractions of all the 141 identified metabolites, in all the 80 samples that were analysed. (b) A circus plot displaying the proportion of each super-class of metabolites in relation to the age or the cocoon shell or the presence/absence of colour. (c) Decision tree analysis47 showing the chemical structure of Fumaric acid and the signal intensity fraction above or below which old or contemporary samples can be distinguished. (d) Validation for the presence of flavonoids in the 148 cocoon shell samples. The results are expressed as rutin equivalents. (e) Validation of the presence of free amino acids in cocoon shells33. A set of 10 contemporary (Upper 10 labels on the Y-axis) and 10 old (Lower 10 labels on the Y-axis) cocoon shell samples were analysed for the presence of 18 free amino acids (X-axis) as identified in the metabolomics analysis (see Fig. 4a). The heatmap shows the content of each amino acid expressed as Log of μg per cocoon shell. No statistically significant differences (p > 0.05) were recorded in the free amino acids contents between the two groups of cocoon shells.
To explore potential discriminating features, Decision Tree analysis was applied47. Fumaric acid content was identified as the metabolite that separated old from contemporary cocoon shell samples (Fig. 4c), with higher levels measured in contemporary shells, and this was the only clear two group classification we identified47. Fumaric acid is a weak acid, an intermediate of the TCA cycle and a known bacterial growth inhibiting compound82,83.
Among the 141 metabolites, three flavonoids were identified: quercetin, quercetin-3,4′-O-di-beta-glucopyranoside and riboflavin44. The fluorescent flavonoids quercetin-5-O-glucoside and quercetin-5,4′-O-glucoside70 were not included in the metabolite database and therefore were not identified, although fluorescence imaging (Fig. 2j) suggested their possible presence.
To validate flavonoid detection, total flavonoid content was quantified using the protocol of Lu et al.12. Cluster 2 samples (Fig. 2k) had the highest measured amounts, while most other clusters contained 10–40 μg flavonoids per 100 mg cocoon shell (Fig. 4d). The highest amount of total flavonoids was measured in the Daizo race (Fig. 4d), an Asian race that carries the + Lg/ + Lg genotype and thereby accumulates prolinylflavonols on its cocoon in addition to other flavonoids84. On the other hand, the lowest amount of flavonoids was measured in the race Baghdad, a race the carries the Yellow Inhibitor (I/I) genotype85 and had just 0.54 μg rutin equivalent of total flavonoids and also had the lowest florescence intensity of all the cocoon shell samples (see Figs. 2j, 4d).
To further validate metabolite detection, free amino acid content was analysed by LC-HR-MS/MS33. All 18 tested amino acids were detected in all 20 samples analysed, supporting the metabolomics pipeline results. Most amino acids were detected in the range of 1–21 μg per cocoon shell, with tryptophan and methionine detected at lower levels of 90 ng and 50 ng per cocoon shell, respectively (Fig. 4e and33). No statistically significant difference (p > 0.05) was observed in free amino acid content between contemporary and old cocoon shells (Fig. 4e).
Lipidomics data validation
LC-MS/MS lipidomics analysis of 80 cocoon shell samples identified 981 distinct lipids46. Most of the lipids were classified as ceramides and phytoceramides, followed by sphingolipids (Fig. 5a and46). To our knowledge, this dataset represents the first lipidomics dataset for B. mori cocoon shells. When the identified lipids were grouped into different super-classes and sub-classes46, a Circos plot showed that ceramides constituted the majority, with phytoceramides as the largest subclass, followed by sphingolipids (Fig. 5a and46).
B. mori cocoon shell lipidomics. (a) A circus plot displaying the proportion of each super-class or sub-class of lipids in relation to the age of the cocoon shell or the presence/absence of colour. (b) Decision tree analysis47 showing the chemical structure of CAR 22:1 and the signal intensity fraction above or below which old or contemporary samples can be distinguished. (c) Validation of the presence of lipids in cocoon shells. A set of 10 contemporary (Black bars) and 10 old (White bars) cocoon shell samples were analysed for the presence of lipids as identified in the lipidomics analysis (see Fig. 5a). Results are expressed as oleic acid equivalents per cocoon shell. Oleic acid had an EC50 of 18.9 μg in this experimental setup (see Inset figure and Methods section). (d) Validation of the presence of phospholipids in cocoon shells. The same as above set of 10 contemporary (Black bars) and 10 old (White bars) cocoon shell samples were analysed for the presence of phospholipids as identified in the lipidomics analysis (see Fig. 5a). Results are expressed as μg of Phosphorus equivalents per cocoon shell. Potassium dihydrogen phosphate had an EC50 of 34.2 μg in this experimental setup (see Inset figure and Methods section).
Decision Tree analysis of the lipidomics dataset indicated47 that the signal intensity fraction of CAR 22:1 relative to the total signal intensity of each sample distinguished old from contemporary cocoon shells (Fig. 5b). CAR 22:1 is an acylcarnitine whose biological role is to transport acyl groups from the cytosol into the mitochondrial matrix for β-oxidation86.
To validate lipid detection independently, two biochemical assays were performed on the same 20 cocoon shell samples that were analysed for free amino acids. Total lipid content was measured using the sulfo-phospho-vanillin protocol39 (Fig. 5c) and phospholipid content was measured using a modified Bartlett’s assay35 (Fig. 5d).
The 20 analysed samples contained 267.5 ± 192.2 μg (Mean ± SD) of oleic acid equivalents on average (Fig. 5c). Phospholipid content averaged 12.5 ± 5.8 μg per cocoon shell, corresponding to approximately 4.6% of the total lipid content (compare Fig. 5c,d). Statistical analysis showed a significant difference in total lipid content between old and contemporary cocoon shells (p = 0.042, unpaired t-test with Welch’s correction), since some old cocoon shells contained relatively high amounts of total lipids, whereas phospholipid content did not differ significantly between the two groups (p = 0.58, unpaired t-test with Welch’s correction).
Clustering analysis validation
All validation results described above did not reveal clear differences between cocoon shell samples, consistent with the selection of 80 out of 148 samples22. However, Decision Tree analysis47 had identified some differences between contemporary and old cocoon shells (Fig. 4c and Fig. 5b). Since we calculated the original RGB and HSB values of the old cocoon shell samples to approximately reconstitute their colour and aiming to assess grouping more systematically, we performed Partial Least Squares–Discriminant Analysis (PLS-DA) analysis40 in MetaboAnalyst 6.0 (https://www.metaboanalyst.ca/), using three independent variables: age (contemporary/old), colour (coloured/white), and sample origin (Asia/Europe). When age was used as the variable, contemporary and old cocoon shells separated clearly in both the metabolomics and lipidomics datasets (Fig. 6a,d). When colour was used as the variable, clusters overlapped substantially (Fig. 6b,e). When origin was used, old samples from the Middle East, Georgia, and Armenia grouped closer to old European samples, while contemporary samples formed a distinct cluster (Fig. 6c,f). The lipid profiles of contemporary samples clustered tightly, with no grouping pattern observed by geographic origin (Italy, Japan, Georgia) (Fig. 6d–f).
B. mori cocoon shell clustering validation. (a) PLS-DA analysis of the identified metabolites using cocoon shell age as independent variable40. The labels next to each circle, for this and all subsequent figures, indicates the samples’ names19. (b) PLS-DA analysis of the identified metabolites using cocoon shell colour (white/coloured) as independent variable40. (c) PLS-DA analysis of the identified metabolites using cocoon shell origin (Asian/European) as independent variable40. (d) PLS-DA analysis of the identified lipids using cocoon shell age as independent variable40. (e) PLS-DA analysis of the identified lipids using cocoon shell colour (white/coloured) as independent variable40. (f) PLS-DA analysis of the identified lipids using cocoon shell origin (Asian/European) as independent variable40. (g) Two-dimensional presentation of the results from the analysis of the normalized signal intensity values of the identified proteins using the Locally Linear Embedding (LLE) algorithm49. A 3D interactive map of the same results can be viewed in50. (h) Two-dimensional presentation of the results from the analysis of the normalized signal intensity values of the identified metabolites using the Locally Linear Embedding (LLE) algorithm49. A 3D interactive map of the same results can be viewed in50. (i) Two-dimensional presentation of the results from the analysis of the normalized signal intensity values of the identified lipids using the Locally Linear Embedding (LLE) algorithm49. A 3D interactive map of the same results can be viewed in50. (j) Hierarchical clustering dendrogram of the cocoon shell samples upon analysis of the normalized signal intensity values of the identified proteins (see Fig. 3a and67). The dotted circle indicates the two samples of cocoon shells of Japanese origin (one old cocoon shell sample and one of a Japanese hybrid strain). The asterisks (*) indicate samples with unusual clustering, i.e. old cocoon shells samples clustering with contemporary ones or vice versa (see19 for detains). (k) Hierarchical clustering dendrogram of the cocoon shell samples upon analysis of the normalized signal intensity values of the identified metabolites (see Fig. 4a and19,67 for detains). (l) Hierarchical clustering dendrogram of the cocoon shell samples upon analysis of the normalized signal intensity values of the identified lipids46,67. (m) Venn diagrams showing the grouping of the 80 cocoon shell samples that were analysed by proteomic, metabolomic and lipidomic analysis upon clustering of the samples using the k-means clustering algorithm. The clustering of the samples when k = 2 is shown68. The left and right Venn diagram differ by the clustering of the proteomics dataset by the k-means algorithm. The two cocoon shell samples on the left image are those samples closest to the centroid point of the clusters and the cocoon shell sample on the right image is the sample closest to the centroid point of the other clusters. (n) The World atlas depicted in Fig. 1a but this time layered with shaded areas and abstract lines that depict the expansion of silkworm races from Asia to Europe and their further reciprocal human-driven migration within Europe based on validation from historical manuscripts. The three indigo dots show the location of the three samples identified in the k-means clustering analyses shown in Fig. 6m.
To address limitations of PLS-DA40,87,88, Locally Linear Embedding (LLE) was applied to proteomics (Fig. 6g), metabolomics (Fig. 6h), and lipidomics (Fig. 6i) datasets. The two-dimensional projections from LLE revealed a continuum among old cocoon shell samples, which merged into the space occupied by contemporary samples (Fig. 6g–i). In the lipidomics dataset, contemporary samples formed a compact cluster, positioned between old samples from the Middle East and China/Indian subcontinent (Fig. 6i). Interactive 3D renderings of these results are available in50.
Hierarchical clustering57 further tested these separations. For proteomics, clusters formed along an old/contemporary divide (Fig. 6j). Within this divide, however, contemporary cocoon shell samples were placed with old cocoon shell samples and vice versa (Fig. 6j). There were two main clusters that segregated the old cocoon shell samples with a crown cluster of mostly samples from the Indian subcontinent (Fig. 6j). Moreover, two Japanese samples, one contemporary and one old, were placed together (Fig. 6j and67). Contemporary cocoon shells also formed a distinct group containing samples from multiple origins (Italy, Georgia, Japan) (Fig. 6j). Hierarchical clustering of metabolomics data similarly grouped contemporary samples together regardless of geographic origin (Japan, Italy, Georgia) (Fig. 6k and67). For lipidomics, hierarchical clustering separated old from contemporary samples and resolved three distinct clusters among the old: a small group from Central Asia, a larger cluster branching from East Asia, and a third cluster further divided into two sub-clusters (Fig. 6l and67).
To identify representative samples across datasets, the k-means clustering algorithm57 provided a numerical means of clustering samples across all our datasets (Fig. 6m). The k-means clustering analysis reached k = 40 for the 80 cocoon shell samples for all datasets68. Using silhouette values57,69, k = 2 was selected as the most robust grouping across all datasets. Venn diagram comparisons of clusters (Fig. 6m) revealed that 56 of the 80 samples (Fig. 6m, left image) were consistently grouped across proteomics cluster 0, metabolomics cluster 1, and lipidomics cluster 1. This cluster contained all contemporary cocoon shells and several old ones. Within it, samples No. 43 (Cosenza, Italy) and No. 61 (Var, France) were closest to the clusters centroids. When proteomics cluster 1 was included instead of cluster 0 (Fig. 6m, right image), sample No. 44 (Iran) was closest to the clusters’ centroids. By cross-examining Fig. 2k and Fig. 6m, we observed that sample No. 44 clusters in Cluster 1 (Fig. 2k), sample No. 43 clusters in Cluster 5 (Fig. 2k) together with many contemporary European races (Fig. 2k) while sample No. 61 clusters in Cluster 3 together with the remaining contemporary European races (Fig. 2k).
Validation through historical texts
The k-means clustering results (Fig. 6m) of the combined proteomics, metabolomics, and lipidomics datasets were cross-checked against information recorded in historical manuscripts. These sources confirm that the geographic origins of the three representative cocoon shell samples - produced in 1895 (Cosenza, Italy), 1959 (Var, France), and circa 1820 (Iran)19 - correspond to regions documented as active centres of silkworm egg trading and silkworm rearing10,15,16,89,90,91. The chronological metadata for these samples19 aligns with written accounts that describe European silkworm races and their distribution across different time periods92,93,94.
Historical manuscripts also document sericultural activity in the specific regions where these cocoon shell samples originated. For example, South Italy is consistently described as a sericultural centre94, while N. Rondot95 noted that races in Calabria were described as originating from Persia. The Var region in South France is documented as part of a broader sericultural area that included North Italy and South France during the 19th and 20th centuries94. Iran is described in early 19th-century sources as a region from which silkworm races were obtained15,89,95.
The clustering of cocoon shell samples shown in Fig. 2k was further cross-referenced with descriptions in 19th-century manuscripts that recorded the presence of European silkworm races, which were often assigned place-specific names and documented alongside Chinese and Japanese races introduced into Europe in the late 19th and early 20th centuries10,15,16,89,90,91,93,94. These textual records provide supporting evidence for the labels attached to both old and contemporary cocoon shell samples in this study.
As an additional validation, historical documentation was used to verify the originality and interpretation of the labels found on old cocoon shell samples. Because several samples carried designations that are no longer readily interpretable to a non-specialist, while in some cases, misattribution in past cataloguing may have occurred22, manuscripts spanning the 15th to the 20th centuries were examined for descriptive and pictorial information relevant to cocoon and silkworm races. Manuscripts from the 15th - 18th centuries94,96,97,98 provided early descriptive accounts, while 19th-century books10,16 and multiple manuscripts89,90,91,93,95,99,100,101 provided detailed descriptions and illustrations that correspond to many of the old cocoon shell samples we analysed. These documents also record exchanges of silkworm races between European and Asian regions during the 19th century, consistent with widespread trading of silkworm eggs (Fig. 6n). Manuscripts from the 20th century9 and research papers from the 21st century3,4,102 were additionally consulted to align the labels19 assigned to old cocoon shell samples with those documented in historical sources and modern databases.
Data availability
The data that support the findings of this study are openly available in three data repositories. In detail, the raw files and annotated data of the metabolomics and lipidomics analyses are available in the Metabolomics Workbench, https://www.metabolomicsworkbench.org59. where they have been assigned Study ID ST003842 and ST003843, respectively. The data can be accessed directly via its Project https://doi.org/10.21228/M88V68.
The mass spectrometry proteomics raw data are publicly available via ProteomeXchange with identifier PXD06235161.
All other data that support the findings of this study are openly available and archived on Figshare (https://figshare.com/) (see Data Records section for individual data record URLs).
Code availability
The custom code for hierarchical clustering and k-means clustering analysis and the custom code for the various dimensionality reduction methods that were used in this study (i.e. t-SNE, UMAP, PCA, MDS, ISOMAP and LLE) are openly available on Figshare57. Briefly, the code, presented in two separate files, consists of custom Python code that can be executed in the Google Colab environment using the datasets of44,45,46 to generate (i) the hierarchical clustering and k-means analyses results and (ii) the 3D plots of the various dimensionality reduction methods.
References
Sun, W. et al. Phylogeny and evolutionary history of the silkworm. Science China Life Sciences 55, 483–496, https://doi.org/10.1007/s11427-012-4334-7 (2012).
Yang, S. Y. et al. Demographic history and gene flow during silkworm domestication. BMC Evol Biol 14, 185, https://doi.org/10.1186/s12862-014-0185-0 (2014).
Tong, X. et al. High-resolution silkworm pan-genome provides genetic insights into artificial selection and ecological adaptation. Nature Communications 13, 5619, https://doi.org/10.1038/s41467-022-33366-x (2022).
Xiang, H. et al. The evolutionary road from wild moth to domestic silkworm. Nat Ecol Evol 2, 1268–1279, https://doi.org/10.1038/s41559-018-0593-4 (2018).
Dozy, R. P. A. Le calendrier de Cordoue de l’année 961: texte arabe et ancienne traduction latine. (E.J. Brill, 1961).
Spiro, F. Pausaniae Graeciae descriptio. (Teubner, 1903).
Ayuzawa, C. et al. Handbook of Silkworm Rearing. (Fuji Publishing Co., 1972).
Aruga, H. Principles of Sericulture. (Taylor & Francis, 1994).
Hiratsuka, E. Silkwrom Breeding. 1st Edition, (Sericulture Science Research Centre/CRC Press, 1969).
Quajat, E. Dei Bozzoli piu pregevoliche preparano i lepidotteri setiferi. (Fratteli Drucker, Verona, Italy, 1904).
Toyama, K. Hyakunen Izen ni Okeru Honpō Kaiko no Shurui. Dai Nihon Sanshi Kaihō 9, 1–9 (1900).
Lu, Y. et al. Deciphering the Genetic Basis of Silkworm Cocoon Colors Provides New Insights into Biological Coloration and Phenotypic Diversification. Mol Biol Evol 40, https://doi.org/10.1093/molbev/msad017 (2023).
Mirhoseini, S. Z., Dalirsefat, S. B. & Pourkheirandish, M. Genetic Characterization of Iranian Native Bombyx mori Strains Using Amplified Fragment Length Polymorphism Markers. Journal of Economic Entomology 100, 939–945, https://doi.org/10.1093/jee/100.3.939 (2007).
Seyf, A. Silk production and trade in Iran in the nineteenth century. Iranian Studies 16, 51–71, https://doi.org/10.1080/00210868308701605 (1983).
Cornalia, E. Monografia del bombice del Gelso (Bombix mori linn.). (Tipografia di Giuseppe Bernardoni di Gio, 1856).
Duseigneur-Kléber, E. Monographie du cocon de soie. (impr. Pitrat, 1862).
Holvast, E. J., Celik, M. A., Phillips, M. J. & Wilson, L. A. B. Do morphometric data improve phylogenetic reconstruction? A systematic review and assessment. BMC Ecology and Evolution 24, 127, https://doi.org/10.1186/s12862-024-02313-3 (2024).
Lee, M. S. Y. & Palci, A. Morphological Phylogenetics in the Genomic Age. Current Biology 25, R922–R929, https://doi.org/10.1016/j.cub.2015.07.009 (2015).
Fragkou, P. et al. Bombyx mori cocoon shell collection descriptor. figshare https://doi.org/10.6084/m9.figshare.29974921 (2025).
Fragkou, P. et al. Phenomics and Biometric data of the 148 cocoon shells samples. figshare https://doi.org/10.6084/m9.figshare.29975173 (2025).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682, https://doi.org/10.1038/nmeth.2019 (2012).
Fragkou, P. et al. Selection justification for cocoon shell omics analyses. figshare https://doi.org/10.6084/m9.figshare.29975101 (2025).
Rackov, N. et al. Bacterial cellulose: Enhancing productivity and material properties through repeated harvest. Biofilm 9, 100276, https://doi.org/10.1016/j.bioflm.2025.100276 (2025).
Tsugawa, H. et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nature Methods 12, 523–526, https://doi.org/10.1038/nmeth.3393 (2015).
Drotleff, B. & Lämmerhofer, M. Guidelines for Selection of Internal Standard-Based Normalization Strategies in Untargeted Lipidomic Profiling by LC-HR-MS/MS. Anal Chem 91, 9836–9843, https://doi.org/10.1021/acs.analchem.9b01505 (2019).
Wadie, B. et al. METASPACE-ML: Context-specific metabolite annotation for imaging mass spectrometry using machine learning. Nat Commun 15, 9110, https://doi.org/10.1038/s41467-024-52213-9 (2024).
Hughes, C. S. et al. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat Protoc 14, 68–85, https://doi.org/10.1038/s41596-018-0082-x (2019).
Demichev, V., Messner, C. B., Vernardis, S. I., Lilley, K. S. & Ralser, M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nature Methods 17, 41–44, https://doi.org/10.1038/s41592-019-0638-x (2020).
Moulos, P., Samiotaki, M., Panayotou, G. & Dedos, S. G. Combinatory annotation of cell membrane receptors and signalling pathways of Bombyx mori prothoracic glands. 3, 160073, https://doi.org/10.1038/sdata.2016.73 (2016).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods 13, 731–740, https://doi.org/10.1038/nmeth.3901 (2016).
Kohler, G. & Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497, https://doi.org/10.1038/256495a0 (1975).
Kotsiri, M. et al. Should I stay or should I go? The settlement-inducing protein complex guides barnacle settlement decisions. The Journal of Experimental Biology 221, jeb185348, https://doi.org/10.1242/jeb.185348 (2018).
Fragkou, P., Thomaidis, N. S., Kostakis, M. G., Barcenas, M. & Dedos, S. G. Free amino acids validation results (Raw Data). figshare https://doi.org/10.6084/m9.figshare.29975932 (2025).
Papastavropoulou, K., Koupa, A., Kritikou, E., Kostakis, M. & Proestos, C. Edible Insects: Benefits and Potential Risk for Consumers and the Food Industry. Biointerface Research in Applied Chemistry 12, 5131–5149, https://doi.org/10.33263/BRIAC124.51315149 (2021).
Bartlett, G. R. Phosphorus assay in column chromatography. J Biol Chem 234, 466–468, https://doi.org/10.1016/S0021-9258(18)70226-3 (1959).
Partovi, S. E. et al. Coenzyme M biosynthesis in bacteria involves phosphate elimination by a functionally distinct member of the aspartase/fumarase superfamily. J Biol Chem 293, 5236–5246, https://doi.org/10.1074/jbc.RA117.001234 (2018).
Rouser, G., Fkeischer, S. & Yamamoto, A. Two dimensional then layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids 5, 494–496, https://doi.org/10.1007/bf02531316 (1970).
Sugai, A., Sakuma, R., Fukuda, I., Itoh, Y. H. & Itoh, T. Improved Method for Determining Soybean Phospholipid Composition by Two-dimensional TLC-phosphorus Assay. Journal of Japan Oil Chemists’ Society 41, 1029–1034, https://doi.org/10.5650/jos1956.41.1029 (1992).
Knight, J. A., Anderson, S. & Rawle, J. M. Chemical basis of the sulfo-phospho-vanillin reaction for estimating total serum lipids. Clin Chem 18, 199–202, https://doi.org/10.1097/00000000-18-7-673 (1972).
Fragkou, P., Martakos, I., Thomaidis, N. S., Barcenas, M. & Dedos, S. G. PLS-DA analysis results for the metabolomics and lipidomics datasets. figshare https://doi.org/10.6084/m9.figshare.29976385 (2025).
Ge, S. X., Jung, D. & Yao, R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629, https://doi.org/10.1093/bioinformatics/btz931 (2019).
Ryan, M. C. et al. Interactive Clustered Heat Map Builder: An easy web-based tool for creating sophisticated clustered heat maps. F1000Res 8, https://doi.org/10.12688/f1000research.20590.2 (2019).
Vélez-Bermúdez, I. C., Lin, W. D., Chou, S. J., Chen, A. P. & Schmidt, W. Transcriptome and translatome comparison of tissues from Arabidopsis thaliana. Sci Data 12, 504, https://doi.org/10.1038/s41597-025-04805-3 (2025).
Fragkou, P. et al. Metabolomics results for 80 Bombyx mori cocoon shell samples. figshare https://doi.org/10.6084/m9.figshare.29975479 (2025).
Fragkou, P. et al. Proteomics results for 81 Bombyx mori cocoon shell samples. figshare https://doi.org/10.6084/m9.figshare.29975422 (2025).
Fragkou, P. et al. Lipidomics results for 80 Bombyx mori cocoon shell samples. figshare https://doi.org/10.6084/m9.figshare.29975560 (2025).
Fragkou, P., Kotsiantis, S., Barcenas, M. & Dedos, S. G. Decision trees analysis results for the proteomics, metabolomics and lipidomics datasets. figshare https://doi.org/10.6084/m9.figshare.29975653 (2025).
Loh, W.-Y. Classification and regression trees. WIREs Data Mining and Knowledge Discovery 1, 14–23, https://doi.org/10.1002/widm.8 (2011).
Fragkou, P., Kotsiantis, S., Barcenas, M. & Dedos, S. G. 2D dimensionality reduction plots for the proteomics, metabolomics and lipidomics datasets. figshare https://doi.org/10.6084/m9.figshare.29976331 (2025).
Fragkou, P., Kotsiantis, S., Barcenas, M. & Dedos, S. G. 3D Interactive dimensionality reduction plots for the proteomics, metabolomics and lipidomics datasets. figshare https://doi.org/10.6084/m9.figshare.27985811 (2025).
Maaten, L. V. D. & Hinton, G. E. Visualizing Data using t-SNE. Journal of Machine Learning Research 9, 2579–2605 (2008).
Healy, J. & McInnes, L. Uniform manifold approximation and projection. Nature Reviews Methods Primers 4, 82, https://doi.org/10.1038/s43586-024-00363-x (2024).
Jia, W., Sun, M., Lian, J. & Hou, S. Feature dimensionality reduction: a review. Complex & Intelligent Systems 8, 2663–2693, https://doi.org/10.1007/s40747-021-00637-x (2022).
Tenenbaum, J. B., Silva, V. D. & Langford, J. C. A Global Geometric Framework for Nonlinear Dimensionality Reduction. Science 290, 2319–2323, https://doi.org/10.1126/science.290.5500.2319 (2000).
Roweis, S. T. & Saul, L. K. Nonlinear Dimensionality Reduction by Locally Linear Embedding. Science 290, 2323–2326, https://doi.org/10.1126/science.290.5500.2323 (2000).
Donoho, D. L. & Grimes, C. Hessian eigenmaps: Locally linear embedding techniques for high-dimensional data. Proceedings of the National Academy of Sciences 100, 5591–5596, https://doi.org/10.1073/pnas.1031596100 (2003).
Fragkou, P., Kotsiantis, S., Barcenas, M. & Dedos, S. G. Custom Google Colab executable codes. figshare https://doi.org/10.6084/m9.figshare.28710992 (2025).
Fragkou, P., Martakos, I., Kotsiantis, S., Barcenas, M. & Dedos, S. G. Raw files and annotated data of the metabolomics and lipidomics analyses of 80 cocoon shell samples. Metabolomics Workbench https://doi.org/10.21228/M88V68 (2025).
Sud, M. et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res 44, D463–470, https://doi.org/10.1093/nar/gkv1042 (2016).
Perez-Riverol, Y. et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res 53, D543–d553, https://doi.org/10.1093/nar/gkae1011 (2025).
Fragkou, P., Rouni, G., Samiotaki, M., Barcenas, M. & Dedos, S. G. Mass spectrometry proteomics raw data of 81 Bombyx mori cocoon shell samples. PRIDE https://www.ebi.ac.uk/pride/archive/projects/PXD062351 (2025).
Fragkou, P. et al. Adjusted absorbance measurements of the 148 cocoon shells samples.xlxs. figshare https://doi.org/10.6084/m9.figshare.29975215 (2025).
Fragkou, P. et al. Fluorescence intensity measurements of 47 cocoons shell samples. figshare https://doi.org/10.6084/m9.figshare.29975242 (2025).
Fragkou, P. et al. Phenomics data of the148 cocoon shells samples (RGB and HSB values). figshare https://doi.org/10.6084/m9.figshare.29975275 (2025).
Fragkou, P. et al. Adjusted phenomics data of the148 cocoon shells samples (Original and adjusted RGB and HSB values). figshare https://doi.org/10.6084/m9.figshare.29975305 (2025).
Fragkou, P. et al. Clustering data of the148 cocoon shells samples based on phenomics analysis. figshare https://doi.org/10.6084/m9.figshare.29975329 (2025).
Fragkou, P., Kotsiantis, S., Barcenas, M. & Dedos, S. G. Hierarchical clustering analysis results for the proteomics, metabolomics and lipidomics datasets. figshare https://doi.org/10.6084/m9.figshare.29976298 (2025).
Fragkou, P., Kotsiantis, S., Barcenas, M. & Dedos, S. G. k-means clustering results of the proteomics, metabolomics and lipidomics datasets. figshare https://doi.org/10.6084/m9.figshare.29975617 (2025).
Fragkou, P., Kotsiantis, S., Barcenas, M. & Dedos, S. G. Silhouette values of the k-means analysis. figshare https://doi.org/10.6084/m9.figshare.29976445 (2025).
Daimon, T. et al. The silkworm Green b locus encodes a quercetin 5-O-glucosyltransferase that produces green cocoons with UV-shielding properties. Proceedings of the National Academy of Sciences 107, 11471–11476, https://doi.org/10.1073/pnas.1000479107 (2010).
Chen, R. et al. Comparative analysis of proteins from Bombyx mori and Antheraea pernyi cocoons for the purpose of silk identification. J Proteomics 209, 103510, https://doi.org/10.1016/j.jprot.2019.103510 (2019).
Lee, B., Pires, E., Pollard, A. M. & McCullagh, J. S. O. Species identification of silks by protein mass spectrometry reveals evidence of wild silk use in antiquity. Scientific Reports 12, 4579, https://doi.org/10.1038/s41598-022-08167-3 (2022).
Liu, C. et al. Osiris9a is a major component of silk fiber in lepidopteran insects. Insect Biochemistry and Molecular Biology 89, 107–115, https://doi.org/10.1016/j.ibmb.2017.09.002 (2017).
Zhang, Y. et al. Comparative Proteome Analysis of Multi-Layer Cocoon of the Silkworm, Bombyx mori. PLOS One 10, e0123403, https://doi.org/10.1371/journal.pone.0123403 (2015).
Guo, X. et al. Proteins in the Cocoon of Silkworm Inhibit the Growth of Beauveria bassiana. PLOS One 11, e0151764, https://doi.org/10.1371/journal.pone.0151764 (2016).
Dong, Z., Xia, Q. & Zhao, P. Antimicrobial components in the cocoon silk of silkworm, Bombyx mori. International Journal of Biological Macromolecules 224, 68–78, https://doi.org/10.1016/j.ijbiomac.2022.10.103 (2023).
Fang, S. M., Zhou, Q. Z., Yu, Q. Y. & Zhang, Z. Genetic and genomic analysis for cocoon yield traits in silkworm. Sci Rep 10, 5682, https://doi.org/10.1038/s41598-020-62507-9 (2020).
Gao, Y. et al. Imaginal disc growth factor maintains cuticle structure and controls melanization in the spot pattern formation of Bombyx mori. PLoS Genet 16, e1008980, https://doi.org/10.1371/journal.pgen.1008980 (2020).
Fahy, E. & Subramaniam, S. RefMet: a reference nomenclature for metabolomics. Nat Methods 17, 1173–1174, https://doi.org/10.1038/s41592-020-01009-y (2020).
Krzywinski, M. I. et al. Circos: An information aesthetic for comparative genomics. Genome Res. 9, 1639–1645, https://doi.org/10.1101/gr.092759.109 (2009).
Garssen, J., Norval, M., Crosby, J., Dortant, P. & Van Loveren, H. The role of urocanic acid in UVB-induced suppression of immunity to Trichinella spiralis infection in the rat. Immunology 96, 298–306, https://doi.org/10.1046/j.1365-2567.1999.00698.x (1999).
Podolak, R. K., Zayas, J. F., Kastner, C. L. & Fung, D. Y. C. Inhibition of Listeria monocytogenes and Escherichia coli O157:H7 on Beef by Application of Organic Acids (†). J Food Prot 59, 370–373, https://doi.org/10.4315/0362-028x-59.4.370 (1996).
Nazir, A., Puthuveettil, A. R., Hussain, F. H. N., Hamed, K. E. & Munawar, N. Endophytic fungi: nature’s solution for antimicrobial resistance and sustainable agriculture. Front Microbiol 15, 1461504, https://doi.org/10.3389/fmicb.2024.1461504 (2024).
Hirayama, C. et al. Deficiency of a pyrroline-5-carboxylate reductase produces the yellowish green cocoon ‘Ryokuken’ of the silkworm, Bombyx mori. Heredity 120, 422–436, https://doi.org/10.1038/s41437-018-0051-8 (2018).
Li, X. et al. Mapping of the yellow inhibitor gene I in silkworm Bombyx mori using SSR markers. Yi Chuan 30, 1039–1042, https://doi.org/10.3724/sp.j.1005.2008.01039 (2008).
Dambrova, M. et al. Acylcarnitines: Nomenclature, Biomarkers, Therapeutic Potential, Drug Targets, and Clinical Trials. Pharmacol Rev 74, 506–551, https://doi.org/10.1124/pharmrev.121.000408 (2022).
Zhvansky, E. et al. Comparison of Dimensionality Reduction Methods in Mass Spectra of Astrocytoma and Glioblastoma Tissues. Mass Spectrom (Tokyo) 10, A0094, https://doi.org/10.5702/massspectrometry.A0094 (2021).
Ruiz-Perez, D., Guan, H., Madhivanan, P., Mathee, K. & Narasimhan, G. So you think you can PLS-DA? BMC Bioinformatics 21, 2, https://doi.org/10.1186/s12859-019-3310-7 (2020).
Clugnet, L. Géographie de la soie: étude géographique et statistique sur la production et le commerce de la soie en cocon. (Georg, 1877).
Silbermann, H. Die Seide, ihre Geschichte, Gewinnung und Verarbeitung…: Die Geschichte der Seidenkultur, des Seidenhandels und der Seidenwebekunst von ihren Anfängen bis auf die Gegenwart. Naturgeschichte der Seide. Die wilden Seiden. Die Gewinnung der Rohseide und Zubereitung der Gespinste. (Verlag von H.A. Ludwig Degener, 1897).
Fillipides, S. Studies on the silkwrom. (Kefalides, N. (eds), 1890).
Ure, A. The Philosophy of Manufactures: Or, An Exposition of the Scientific, Moral, and Commercial Economy of the Factory System of Great Britain. (C. Knight, 1835).
Dandolo, V. C. The Art of rearing Silk-worms. Translated from the work of Count Dandolo. (London, 1825).
Betti, Z. Del baco da seta: canti IV, con annotazione. (Per Antonio Andreoni, 1756).
Rondot, N. L’art de la soie: Les soies. (Imprimerie nationale, 1887).
Vida, M. H. Marci Hieronymi Vidae… De arte poetica lib. III; eiusdem De bombyce lib. II; eiusdem De ludo scacchorum lib. I; eiusdem Hymni; eiusdem Bucolica. (apud Ludouicum Vicentinum, 1527).
Spoleto, G. D. De Sere, seu de setivomis animalibus. (1505).
Lazzarelli, L. Ludovici Lazzarelli Septempedani… Bombyx accesserunt ipsius aliorumque poetarum carmina cum commentariis de vitis eorumdem Joanne Francisco Lancillottio a Staphylo auctore. (apud Petrum Paulum Bonelli, 1765).
Hutton, T. XIV.: On the Reversion and Restoration of the Silkworm (Part II.); with Distinctive Characters of Eighteen Species of Silk-producing Bombycidæ. Transactions of the Royal Entomological Society of London 12, 295–331, https://doi.org/10.1111/j.1365-2311.1864.tb00108.x (1864).
Moore, F. XXIV. On the Asiatic Silk-producing Moths. Transactions of the Royal Entomological Society of London 11, 313–322, https://doi.org/10.1111/j.1365-2311.1862.tb01281.x (1862).
de Bavier, E. La sériciculture, le commerce des soies et des graines et l’industrie de la soie au Japon. (H. Georg, 1874).
Kosegawa, E., Reddy, G. V., Shimizu, K. & Okajima, T. Induction of non-diapause egg by dark and low temperature incubation in local variety of the silkworm, Bombyx mori. The Journal of Sericultural Science of Japan 69, 369–375, https://doi.org/10.11416/kontyushigen1930.69.369 (2000).
Acknowledgements
B. mori cocoon shell samples used in this study were provided by: Mrs. Nino Kuprava, Mrs. Salome Phachuashvili and Mrs. Marina Gonashvili of the State Silk Museum, Tbilisi, Georgia; Dr. Nargiz Baramidze Scientific-Research Base of the Ministry of Agriculture, Sericulture Laboratory, Mtskheta Municipality, Georgia; Dr. Roberto Bruni, Mr. Giuliano De Angelis and Dr. Rosanna Moretti of the Agricultural Institute of Ascoli Piceno, Marche, Italy; Dr. Ana Pagán Bernabeu and Dr. Salvador David Aznar Cervantes Biotechnology, Genomics and Plant Improvement Department, Instituto Murciano de Investigación y Desarrollo Agrario y Ambiental (IMIDA), Murcia, Spain; Dr. Tsuguru Fujii and Dr. Toskiaki Fujimoto, of the Laboratory of Silkworm Genetic Resources, Institute of Genetic Resources, Kyushu University Graduate School of BioResources and Bioenvironmental Science, Fukuoka, Japan; Dr. Eichi Kosegawa and Dr. Eiji Okada,National Agriculture and Food Research Organization (NARO) Genetic Resources Center, Tsukuba, Ibaraki, Japan; Dr. Antonios Tsagkarakis, Laboratory of Sericulture and Apiculture, Agricultural University of Athens, Athens, Greece; Dr Alessandro Giusti Department of Life Sciences, The Natural History Museum, London, UK. The B. mori cocoon shell samples donated by Kyushu University staff were within the framework of the National Bio-Resource Project (NBRP) of MEXT, Japan. The NIH Common Fund’s National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench where the metabolomics and lipidomics raw files have been deposited, is supported by NIH grant U2C-DK119886 and OT2-OD030544 grants. The authors thank Miss Evangelia Moustani for the artwork and design of Fig. 6n. The corresponding author thanks Dr. Claudio Zanier, University of Pisa, Pisa, Italy, for his inspiring guidance on the complex history of silk in Europe. This research was funded by the project: Advocating the Role of silk Art and Cultural heritage at National and European scale - Aracne (European Union’s Horizon Europe Research and Innovation Programme: Grant Agreement No 101095188). We acknowledge support of this work by the project “The Greek Research Infrastructure for Personalised Medicine (pMED-GR)” (MIS 5002802) which is implemented under the Action “Reinforcement of the Research and Innovation Infrastructure”, funded by the Operational Programme & “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014–2020) and co-financed by Greece and the European Union (European Regional Development Fund).
Author information
Authors and Affiliations
Contributions
P.F.: Phenomics analysis of cocoon shells, validation, project management, manuscript preparation; I.M.: Post-acquisition statistical analysis of metabolomics and lipidomics data; G.R. Sample preparation for proteomics analysis; D.V. Monoclonal antibody generation and quality assessment;. E.K.: Phenomics analysis of cocoon shells; A.S. and S.C.: cocoon shell samples’ management and curation; N.S.T: Resources, reagents and instrumentation; M.G.K.: Amino acids analysis and validation; M.S. Proteomics data acquisition and analysis S.K. Mathematical modelling of data; M.B.: Metabolomics and Lipidomics data acquisition and analysis; S.G.D. Project management, data analysis manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fragkou, P., Martakos, I., Rouni, G. et al. Multiomics analysis of the Silkworm cocoon shell. Sci Data 12, 1630 (2025). https://doi.org/10.1038/s41597-025-06071-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-06071-9