Abstract
White Spot syndrome virus (WSSV) causes rapid shrimp mortality and production loss worldwide. This study demonstrates potential use of Lactobacillus johnsonii KD1 as an anti-WSSV agent for post larva shrimp cultivation and explores some potential mechanisms behind the anti-WSSV properties. Treatment of Penaeus vannamei shrimps with L. johnsonii KD1 prior to oral challenge with WSSV-infected tissues showed a significantly reduced mortality. In addition, WSSV copy numbers were not detected and shrimp immune genes were upregulated. Genomic analysis of L. johnsonii KD1 based on Illumina and Nanopore platforms revealed a 1.87 Mb chromosome and one 15.4 Kb plasmid. Only one antimicrobial resistance gene (ermB) in the chromosome was identified. Phylogenetic analysis comparing L. johnsonii KD1 to other L. johnsonii isolates revealed that L. johnsonii KD1 is closely related to L. johnsonii GHZ10a isolated from wild pigs. Interestingly, L. johnsonii KD1 contains isolate-specific genes such as genes involved in a type I restriction-modification system and CAZymes belonging to the GT8 family. Furthermore, genes coding for probiotic survival and potential antimicrobial/anti-viral metabolites such as a homolog of the bacteriocin helveticin-J were found. Protein–protein docking modelling suggests the helveticin-J homolog may be able to block VP28–PmRab7 interactions and interrupt WSSV infection.
Similar content being viewed by others
Introduction
To date the trend of global shrimp production has risen from less than 75,000 metric tons to more than 5.5 million metric tons during 1980–2017 and increased continuously1. Especially Penaeus vannamei (White leg shrimp) and Penaeus monodon (Giant tiger prawn) are the most prominent for shrimp marketing. In Asia, P. vannamei is more cultivated than P. monodon due to specific pathogen-free broodstock, low-cost farming, and providing higher meat yields2. Likewise, Thailand showed production of P. vannamei and P. monodon at total 263,245 and 16,292 tons, respectively in 20143. However, shrimp production is damaged by shrimp pathogens especially white spot syndrome virus (WSSV) which is a double stranded DNA virus and causing concern in Thailand due to broad infection of all penaeid species, rapid dissemination, and causing shrimp mortality 3–10 days after viral infection. Shrimps infected with WSSV show signs of symptoms including white spots, lost cuticle, reduction of feeding, and exhaustion leading to death4. This led to 15% loss of worldwide shrimp production and economic loss at US$ 1 billion5,6.
To reduce WSSV infection during shrimp cultivation, probiotics can be used to promote shrimp growth and protect against pathogens, reducing the use of antibiotics. Lactic acid bacteria (LAB) are Gram-positive and non-spore forming bacteria. They are a highly diverse and heterogenous group containing many species consisting of the genera Lactobacillus, Enterococcus, Lactococcus, Pediococcus, Streptococcus, Leuconostoc, Carnobacterium, and Weissella7,8. Some are also generally recognized as safe (GRAS) for use in humans and animals as probiotics. LAB can activate host immune systems, improve human and animals gastrointestinal (GI) tract immunity, and produce beneficial metabolites such as lactic acid which is an end product of carbohydrate metabolism, can inhibit pathogenic growth and can be applied to food production in humans and animals9.
Lactobacillus johnsonii (L. johnsonii) is a Gram positive, non-spore forming, non-motile, and facultative anaerobic bacteria10. Previously, L. johnsonii has been observed to decrease pathogenic infection and colonization, modulate host immune systems, and improve growth of animals such as pigs and poultries11,12,13,14,15,16. In aquaculture, L. johnsonii was found in the intestine of Dicentrarchus labrax (European sea bass) suggesting the ability of L. johnsonii to colonize in an aquatic animal8,17. However, the probiotic activities of L. johnsonii in aquaculture are not prevalent or clear. Thus, this study screened the role of the probiotic L. johnsonii KD1 in shrimp protection against WSSV and investigated shrimp immune activation. Additionally, the probiotics genome was examined to find genes potentially involved in promoting shrimp survival and pathogen inhibition for application in feed supplementation.
Results and discussion
Evaluation of probiotic bacteria against WSSV
Shrimps submerged with 100X L. johnsonii KD1 were significantly protected against WSSV, starting at 3 days post infection (dpi.) and more evident at 4 dpi., as shrimp mortality was lower than that of shrimps receiving 1X L. johnsonii KD1 and shrimps not receiving any probiotics (positive control) (Fig. 1A). This was consistent with the viral load assessed by qPCR on 100 ng total DNA extracted from the tissues of survived shrimps where WSSV copies were undetectable (Fig. 1B). At 5 dpi., 100X L. johnsonii KD1-treated shrimps showed the lowest shrimp cumulative mortality (38%) when comparing to 1X L. johnsonii KD1-treated shrimps (64%) and positive control (97%), respectively (Fig. 1A). Our findings suggest that a high dose of L. johnsonii KD1 promoted shrimp survival after WSSV infection and the intrinsic mechanism involved in its protection efficacy remains to be explored. Similarly, P. vannamei treated with the same high doses of L. lactis and L. plantarum could delay shrimp mortality and reduced viral progeny after WSSV infection6. Additionally, consortium of probiotics Pediococcus pentosaceus and Staphylococcus hemolyticus on commercial feed fed to P. vannamei could decrease WSSV prevalence to less than 20%18. Other probiotics such as Bacillus megaterium and Bacillus PC465 used in mixing feed were reported to resist WSSV infection in P. vannamei4. This evidence supports the ability of probiotics to inhibit WSSV infection and replication.
Shrimp cumulative mortality after WSSV infection (A) Mean of percentage cumulative mortality ± standard error in shrimps submerged with 4.5 × 108 [1X] CFU/2 L seawater and 4.5 × 1010 [100X] CFU/2 L seawater of L. johnsonii KD1 for five days with n = 30 shrimps per replicate (three replicates/dose/group) while the negative and positive groups without probiotic treatments with n = 30 shrimps per replicate (three replicates/group) followed by WSSV infection except the negative group. (B) Mean of log WSSV copies ± standard error with 3 survived shrimps per group sampled at 3-days post infection (3 dpi.). Corresponding dpi. at p < 0.05 based on ANOVA was tested by Duncan’s test represented by small alphabets (a, b, and c). Asterisks (*) indicate significant differences between the treatment and the positive control on corresponding dpi. at p < 0.05 based on independent sample T-test and Mann–Whitney Test. Limit of detection (LOD).
Shrimp immune system activation by L. johnsonii KD1
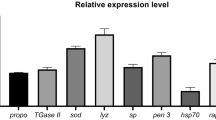
To assess L. johnsonii KD1 ability induce shrimp immune system, we selected four shrimp innate immune genes i.e., genes encoding peroxinectin (PX), prophenoloxidase-1 (proPO I), serine protease (SP), and anti-lipopolysaccharide factor-1 (ALF-1) and determined shrimp gene expression at day 6 (1-day post-feeding) and day 8 (3 days post-feeding) (Fig. 2). In this study, shrimps’ regimen with 100X L. johnsonii KD1 showed upregulation of PX at 1.6 and 2.5 folds on day 6 and day 8, respectively. Consistently, proPO I expression was increased on both days at 2.1 and 1.8 folds, respectively. SP and ALF-1 expression levels were upregulated on day 8 at 2.7 folds and 1.5 folds, respectively.
Shrimp immune gene expression of peroxinectin (PX), serine protease (SP), prophenoloxidase-1 (proPO I), and anti-lipopolysaccharide factor-1 (ALF-1). Shrimp’s regimen with 4.5 × 1010 [100X] CFU/2 L seawater of L. johnsonii KD1 for five days with n = 30 shrimps per replicate (two replicates/group) while the negative group without probiotic treatments with n = 30 shrimps per replicate (two replicates/group). Mean of fold difference of ± standard error with n = 4 shrimps per group at day 6 (or one day post-probiotics regimen in treated groups) and day 8 (or three days post-probiotics regimen). Fold difference of 1 represented as dotted line indicates no change in shrimp gene expression in probiotic treatment in comparison to without probiotic treatment (negative).
In shrimp innate immune response, PX is involved in pathogen recognition, adhesion, and eventually killing6 while proPO I is activated to phenoloxidase-1 by SPs and proPO-activating enzymes (PPAEs) leading to melanin production and pathogen killing19. Previous study reported that both PX and proPO were upregulated when P. vannamei was treated with L. plantarum in mixed feed (104 and 107 CFU/g feed) for 7 days20. Several SPs can also activate PPAEs eventually leading to proPO I induction21. Thus, upregulation of PX, proPO I, and SP in shrimps receiving 100X L. johnsonii KD1 may potentially eliminate WSSV leading to reduced viral load and infection in exposed shrimps. For ALF-1 expression, previous study showed that Marsupenaeus japonicus (kuruma shrimp) treated with L. lactis D1813 in mixed feed (105 CFU/g feed) for 7 days exhibited an upregulation after 3 days and 6 days post-feeding22. ALF-1 is an antimicrobial peptide produced through the Toll pathway in penaeid shrimp. ALF-1 can bind WSSV proteins and block WSSV infection23. Thus, upregulation of ALF-1 expression in 100X L. johnsonii KD1-treated shrimps may potentially prevent WSSV infection.
Genomic characteristics of L. johnsonii KD1
The complete genome of L. johnsonii KD1 was found to consist of one 1,873,159 bp (approximately 1.87 Mb) chromosome named KD1 and one 15,425 bp plasmid named pLJKD1 with 35% and 34% GC content, respectively (Fig. 3). BLASTn of the plasmid exhibited 100% identity to L. johnsonii GHZ10a’s plasmid, and 94% identity to plasmids pUMNLJ21 of L. johnsonii UMNLJ21, pUMNLJ22 of L. johnsonii UMNLJ22, and pLJPF01S of L. johnsonii pf01, respectively. L. johnsonii KD1 contains 1,783 coding sequence (CDS) genes consisting of 1,390 functional genes and 393 genes encoding hypothetical proteins. Additionally, 12 genes encoding rRNA and 74 genes encoding tRNA were found. Eighty-seven genes encoding repeat regions and 9 transporter genes were identified and summarized in Table 1. For plasmid pLJKD1, there are 21 CDS genes consisting of 10 functional genes and 11 genes encoding hypothetical proteins (Table 1, Supplementary Table 1). Genes of the L. johnsonii KD1 chromosome and plasmid were classified into subsystems using the PATRIC database with the main subsystems of protein processing, metabolism, DNA processing and energy identified in the chromosome (Supplementary Fig. 1A) and the main subsystems of energy and stress response, defense and virulence in the probiotic plasmid (Supplementary Fig. 1B). Similarly, among L. johnsonii strains with probiotic activities of pathogenic inhibition and modulation of immune systems in animals such as FI9785, N6.2, ZLJ010, and BS15 showed genomic sizes of 1.76–2.11 Mb with GC content at 35% and CDS genes ranged from 1,718 to 1,959 genes24.
Identification of an antimicrobial resistance (AMR) gene
The AMR gene ermB (99–100% identity) was identified at position 1,623,992–1,624,729 in the probiotic chromosome by ResFinder-4.1 (Table 2). The genes ermT, ermA, ermB, ermY, erm46, and ermR producing the Erm 23S ribosomal RNA methyltransferase have been reported to cause resistance to lincosamide, macrolide, and streptogramin b antibiotics25. Especially, ermB is prevalent in Lactobacillus and confers erythromycin resistance26. This corresponds with another study that identified ermB dependent erythromycin resistance in L. johnsonii27,28. Thus, antibiotic resistant profiles of L. johnsonii KD1 should be further investigated to confirm AMR phenotype according to European Food Safety Authority (EFSA) guideline for safety assessment for probiotic application in feed additives.
Mobile genetic elements (MGEs) and pathogenicity
To validate prophages, PHASTER was used for analysis and showed three regions of prophages in the probiotic chromosome (Table 3, Supplementary Table 2). Coding sequences of prophages showed genes encoding phage proteins, transposases, N-acetylglucosaminyltransferases, a methyltransferase, a phosphoesterase, a hydrolase, transcriptional regulators, an ABC transporter, and hypothetical proteins. Likewise, 24 insertion sequences and 5 composite transposons without association to antimicrobial resistance and virulence factors were identified by MGEfinder (Table 3, Supplementary Table 3). This suggests there will be little dissemination of antibiotic resistance and virulence factors into the environment by L. johnsonii KD1 mobile genetic elements. Additionally, prediction of pathogenicity by PathogenFinder 1.1 showed no pathogenic genes. L. johnsonii KD1 is predicted as a non-human pathogen.
Identification and phylogenetic analysis of L. johnsonii KD1
As the isolation source of L. johnsonii KD1 was unknown, we confirmed from rMLST analysis that the probiotic shares 98% identity with L. johnsonii (Supplementary Table 4). We then proceeded with phylogenetic analysis based on 500 single-copy genes by comparing L. johnsonii KD1 to 21 other L. johnsonii isolates in the NCBI database. The results showed that L. johnsonii KD1 was more closely related to L. johnsonii GHZ10a isolated from Sus scrofa or common wild pig and L. johnsonii ZLJ010 isolated from Sow feces24 (Fig. 4). We could only best suggest that L. johnsonii KD1 could come from swine origin.
Comparative genomic analysis
Comparison of the genomes of L. johnsonii KD1 and 21 L. johnsonii strains identified the pan genome of 4045 genes consisting of 1065 core genes, 1932 accessory genes and 1048 strain specific genes. L. johnsonii KD1 showed nine unique genes encoding for late competent protein COMEA (a DNA receptor), an uncharacterized deacetylase, an integrase, hypothetical proteins, and proteins belonging to the type I restriction-modification system (Supplementary Table 5). Restriction-modification (RM) systems are part of bacterial innate immune systems and work to protect host DNA by preventing cleavage via DNA methylation and to cleave foreign DNA by the action of restriction nucleases (REases)29,30,31. They are classified into type I-IV RM systems, a type I RM system contains (1) a specificity subunit S to recognize a specific DNA motif, (2) two methyltransferase subunit M to methylate DNA, and (3) two restriction endonuclease subunit R to cleave unmethylated DNA. This type cleaves specific sequences far away from a recognition site whereas type II and IV RM systems cleave at a recognition site. Type III RM system cleaves DNA sequences similarly but lack a specificity subunit S29.
Previously, L. johnsonii DPC6026 presented a type III RM system and CRISPR elements that provided phage resistance32. In this study, we found genes coding for a type I RM system (Supplementary Table 6) and one CRISPR element consisting of three spaces and four direct repeats in L. johnsonii KD1 (Supplementary Table 7). This therefore suggests that L. johnsonii KD1 may exhibit phage resistant properties, however, additional experiments are required to investigate this. Due to the fact that phage contamination can be found in food and feed fermentation in non-sterile environments and could lead to low quality and loss of production33, using phage-resistant probiotics could bypass the problems and benefit overall food and feed productions.
As L. johnsonii KD1 in this study provided better anti-WSSV activity than L. plantarum ATCC 14917 according to a previous study6 (Supplementary Fig. 2), genes of L. johnsonii KD1 were compared to two genomes of L. plantarum. The comparative pathways tool identified 11 L. johnsonii KD1 specific pathways classified into 4 classes: xenobiotics biodegradation and metabolism, lipid metabolism, biosynthesis of secondary metabolites and biosynthesis of polyketides and non-ribosomal peptides. The pathways identified were incomplete, with only one or two enzymes present. However, some of products of the enzymatic reactions identified in the pathways have potential anti-viral properties. These are: Glycolate (acetic acid), 4′-hydroxyacetophenone (anti-hepatitis B activity34), 13-OXDE (analogue of Linoleic acid, which is active against WSSV35), and Taxol (HIV-1 pseudovirus inhibition36), (Supplementary Table 8). Then, we found 582 unique genes in L. johnsonii KD1 such as genes encoding type I restriction modification systems, levansucrase, a bacteriocin ABC-transporter with ATP-binding and permease component, and a bacteriocin ABC-transporter with auxiliary protein. However, 268 of the unique genes in L. johnsonii KD1 were identified as hypothetical. To assign function to these proteins the command-line tool MicrobeAnnotator was utilised. This annotated 44 of the 268 unique genes involved in cell wall biosynthesis, glycosylation and lipopolysaccharide biosynthesis, a variety of transporters and 16 viral proteins (Supplementary Table 9). This suggests three possibilities; (1) the proteins annotated as viral proteins share homology with viral proteins but do not share the same function; (2) the probiotic has acquired these proteins and potentially utilises them to carry out functions in the cell or 3) the strain presents viral envelope proteins on their surface as a method of hosting an immune response against the virus. This therefore suggests that different proteins/metabolites may be produced by L. johnsonii KD1 compared to L. plantarum ATCC 14917, which may provide shrimp with a higher protection to WSSV.
Furthermore, to investigate potential differences in carbohydrate usage between the two strains, the carbohydrate active enzymes (CAZymes) were annotated using the dbCAN 2 webserver37. The genes can be classified as belonging to either glycoside hydrolases (GHs), glycosyltransferases (GTs), polysaccharide lyases (PLs), carbohydrate esterases (CEs), auxiliary activities (AAs) and carbohydrate-binding modules (CBMs). The classes can be further categorized based on amino acid sequence homology. As seen in Fig. 5, a variety of different families of CAZymes were identified and both L. plantarum ATCC 14917 genomes had identical profiles. However, when comparing L. johnsonii KD1 and L. plantarum ATCC 14917 there were differences in the relative abundances and families identified. In addition, L. plantarum genomes each contained twofold as many CAZymes as L. johnsonii KD1 (99 and 50 respectively). Interestingly, 10% of L. johnsonii’s CAZymes belonged to the GT8 family (6% GT8, 4% GT8 + GT101), whereas none were identified in L. plantarum ATCC 14917 (Fig. 5A,B). Activities associated with GT8s are lipopolysaccharide, inositol, homogalacturonan, UDP-GlcA: xylan and UDP-Gal:glucoside glycosyltransferases. Another application of the dbCAN2 webserver annotation is the identification of CAZyme gene clusters (CGC’s) which are clusters of genes which work together to digest or utilise carbohydrates38. One of the gene clusters identified in L. johnsonii KD1 contained the two GT8 + GT101 genes and 6 transporter genes, which could present a method of lipopolysaccharide biosynthesis and export (Fig. 5C). The differences observed between the CAZymes profiles of L. johnsonii KD1 and L. plantarum ATCC 14917 genomes demonstrates a difference in carbohydrate usage between the two strains. Furthermore, the appearance of GT8s only in L. johnsonii KD1 suggest this class of enzyme could be important in exerting the probiotic affect observed. In addition, as the GT8 family contains enzymes associated with lipopolysaccharide biosynthesis, the surface of L. johnsonii KD1 may display different lipopolysaccharides to L. plantarum ATCC 14917, which may have probiotic properties39. Moreover, the putative gene cluster (Fig. 5C) identified in the genome of L. johnsonii KD1 suggests a potential carbohydrate modification and export mechanism. Further work could be carried out investigating this pathway and its potential link to the probiotic influence of the strain.
CAZymes profiles. The L. johnsonii KD1 and two L. plantarum ATCC 14917 genomes were annotated via the dbcan2 webserver. Heatmaps of (A) the total number of genes annotated as CAZymes in their respective families and (B) the percentage of each family in respect to total number of CAZymes identified in each genome were produced. (C) GT8 + GT101 gene containing CGC identified in the L. johnsonii KD1 genome (graphic generated by the dbCAN2 webserver); transporter (green), CAZyme (red), and non-signature containing/unannotated genes (grey).
Probiotic survival in shrimp water cultivation and gastrointestinal tract (GI) condition
Probiotics are exposed to salt stress in the water during feeding and by bile salts in the shrimp GI produced by the shrimp hepatopancreas, which can be fatal to some bacteria. Previously, probiotics, especially LABs, have been seen to resist salinity and bile salts40. Accordingly, the genomes of the salt resistant strains of L. plantarum (D31 and T9) contained salt resistant genes such as genes coding for sodium/proton (Na+/H+) antiporters responsible for regulating intracellular pH by sodium efflux out of the cell, a potassium transport system protein, Kup, involved in ion homeostasis during hyperosmotic stress and the transcription factor LysR and RNA polymerase sigma factor RpoD involved in intracellular metabolism homeostasis during salt stress41. In this study, four genes encoding Na+:H+ antiporters, two genes encoding two potassium transport system proteins Kup and two transcription factors; one from the LysR family and the RNA polymerase sigma factor RpoD were identified in L. johnsonii KD1 (Supplementary Table 10). Thus, presenting mechanisms of L. johnsonii KD1 resistance to salt stress. Furthermore, L. johnsonii KD1 revealed three genes coding for choloylglycine hydrolases or bile salt hydrolases (Supplementary Table 10). Bile salt hydrolases deconjugate bile salts leading to release of an amino acid group from steroid core, which has been demonstrated to promote bacterial colonization and survival in the GI tract42. For example, L. acidophilus NCFM contains genes coding for bile salt hydrolases and exhibited bile salt hydrolase activity43. Likewise, L. johnsonii ZLJ010 and L. johnsonii CNCM I-4884 showed genes coding for the Na+:H+ antiporters and choloylglycine hydrolases. Especially, L. johnsonii CNCM I-4884 presented strong bile salt hydrolase activity24,44.
Additionally, heat shock proteins and Clp proteases can repair and refold damaged proteins in a response to stress. Moreover, the UvrABC system protein B and ATP-dependent DNA helicase UvrD/PcrA have been reported to be involved in DNA repair4,45. From this study, the probiotic L. johnsonii KD1’s genome showed genes coding for heat shock proteins, Clp proteases, and DNA repair (Supplementary Table 10). Corresponding to previous studies that reported that L. johnsonii ZLJ010 and CNCMI-4884 genomes contained genes encoding heat shock proteins GroEL, DnaK, and DnaJ, and genes encoding ATP-dependent intracellular proteases ClpP24,44. This suggests that these genes promote L. johnsonii KD1 survival in shrimp ponds and shrimp gut colonization.
Another important factor for probiotics is the ability to colonise the gut of the host. This is impart mediated by the ability of the organism to adhere to the surface of gut cells. Furthermore, a possible probiotic anti-viral mechanism is via interfering with viral attachment and entry by competing for cell binding sites46. Over recent years some of the cell-surface proteins required for this adhesion have been assessed, thus allowing for a list of cell-adhesion proteins identified in Lactobacillus species to be generated from previous studies47,48. The presence of these proteins/homologs in L. johnsonii KD1 and the two L. plantarum genomes was then assessed using the BV-BRC BLASTp tool. A heatmap showing the bit-score value of the top hit generated by the BLASTp was produced (Fig. 6). From the cell-adhesion proteins tested, L. plantarum strains contain more cell adhesion proteins, thus suggesting that these L. plantarum strains have a higher ability to adhere to the cells of the gut and that the antiviral properties of L. johnsonii KD1 are not due to the better ability of the strain to adhere to the cells of the gut to physically block viral infection. However, only a selection of adhesion proteins was tested in this experiment, meaning L. johnsonii KD1 may contain other adhesion proteins not identified here. Future experiments could be carried out to explore the affinity of the different strains to gut cell types and localisation/surface coverage of each strain in the gut of the shrimp. This would allow for a better understanding of each strains ability to adhere to the gut of shrimp and its potential link to the antiviral probiotic affect exhibited by L. johnsonii KD1.
Cell-adhesion protein heat map. A heat map of cell adhesion proteins identified in the genomes of L. johnsonii KD1 and two L. plantarum ATCC 14917 was generated using the BLASTp bit-score value for the top alignment generated for each protein. Proteins above the line belong to mucus/collagen/fibronectin binding proteins. Proteins below the line belong to moonlighting cell adhesion proteins.
Metabolites with potential anti-viral activities
Previously, metabolites such as lactic acid, exopolysaccharides, and antimicrobial compounds have been shown to inhibit viruses49. Lactic acid, a final product of carbohydrate metabolism, plays a role in the innate immune systems of, and is present in all, Lactobacillus. Previous studies reported that lactic acid isomer D and L could inhibit human immunodeficiency virus type 1 (HIV-1) and human simplex virus type 2 (HSV-2) replication50,51,52,53. In this study, we found genes encoding d-lactate dehydrogenase, responsible for d-lactate production and l-lactate dehydrogenase, responsible for l-lactate production (Supplementary Table 10). Both these isomers are the conjugate base of lactic acid, suggesting, the probiotic L. johnsonii KD1 can produce d-lactate and l-lactate that may potentially inhibit WSSV.
Secondly, LAB-produced exopolysaccharides (EPSs) are natural polymers of sugars/ carbohydrates and are involved in adhesion to the host GI tract for colonization, biofilm formation, and antiviral activities such as inhibition of viral infection and replication in Human Adenovirus Type 5 and rotavirus24,49,54,55. In this study, we performed BLASTn of the L. johnsonii KD1 genome using the eps genes of L. johnsonii FI9785 as queries. This identified the core eps gene cluster of epsABCDE involved in EPS biosynthesis, consisting of LytR-transcriptional regulator, polymerization and chain length determination protein, tyrosine kinase, protein-tyrosine-phosphate phosphohydrolase, and undecaprenyl-phosphate galactosephospho-transferase, respectively. Likewise, the glf gene encoding UDP-galactopyranase mutase, epsU gene encoding oligosaccharide translocase, and five genes encoding glycosyltransferases were found (Supplementary Table 10). These were similar to eps gene clusters found in L. johnsonii FI978556,57. This suggests that L. johnsonii KD1’s ability to produce exopolysaccharides could be a potential mechanism for inhibition of WSSV infection. Moreover, inulin is a type of EPSs that increased PO activity and reduced WSSV prevalence in P. vannamei58. Additionally, inulin increased shrimp immune response and changed gut microbiota59. Inulin and levan are produced by inulosucrase and levansucrase enzymes, respectively, with both enzymes belonging to the fructosyltransferases family60. We found that L. johnsonii KD1 contains a gene coding for fructosyltransferase that was annotated as levansucrase at the position of 1,288,765–1,291,003 (Supplementary Table 10). However, L. johnsonii NCC 533 contains a fructosyltransferase (ftf) gene annotated as levansucrase which was able to produce inulin60,61. Thus, we performed a BLASTp of this protein against the UniProt database which showed a 72.5% identity (E-value = 0) hit to levansucrase and a 54.6% identity (E value = 0) to inulosucrase. This suggests that L. johnsonii KD1 could produce levan or inulin which could inhibit WSSV.
Thirdly, antimicrobial peptides such as bacteriocins have been demonstrated to inhibit viral infection, replication and modulate the host immune system49,62. To assess whether a potential mechanism by which L. johnsonii KD1 exerts its antiviral property is through the expression of a natural product, two natural product identification tools, AntiSMASH and BEGEL4, were utilized to analyse the genomes of L. johnsonii KD1 and L. plantarum ATCC 14917. This identified, in L. johnsonii KD1, a homolog of the bacteriocin helveticin-J originally produced by Lactobacillus helveticus 481 and a biosynthetic gene cluster (BGC) in which 55% of the genes had similarity to the BGC of gassericin T (Fig. 7). However, the cluster containing homology to gassericin T lacked the presence of the active peptides GatA and GatX. In addition, BLASTp analysis was unable to identify GatA or GatX homologs in the genome of L. johnsonii KD1. Further analysis of downstream genes identified a mobile genetic element, two bacteriocin immunity proteins and a homolog to holin. This is interesting as holins are encoded by bacteriophages to form pores in the membranes of bacteria enabling the release of endolysins to degrade the cell wall and induce death. However, studies have shown that exogenous application of holin and an endolysin from a phage called SMP can lyse pathogens such as Staphylococcus aureus and Streptococcus suis63. This could potentially allow for a mechanism of competing with other microbes in the absence of GatA/GatX.
Biosynthetic gene clusters identified by antiSMASH and BAGEL4. (A) Bacteriocin helveticin-J homolog and gene neighbourhood identified by BAGEL4. (B) Holin containing gassericin T homolog cluster identified by antiSMASH. (C) gassericin T cluster from Lactobacillus gasseri. Gene annotations were produced by either antiSMASH or BAGEL4 plus further analysis using NCBI BLASTp, UniProt BLASTp and comparison to the BV-BRC annotation of L. johnsonii KD1's genome.
BAGEL4 predicted the presence of a 352 amino acid protein, with 44.5% sequence similarity to the known bacteriocin helveticin-J. However, BLASTp of the predicted sequence against L. johnsonii KD1s genome identified a hit with 94% homology to the query, with the BAGEL4 prediction containing a 21 amino acid extension at the N-terminus. This is due to the BAGEL4 software predicting the start codon to be TTG (Leucine), whereas the BV-BRC RASTtk predicted the sequence to start from the downstream ATG (Methionine). Furthermore, downstream of the helveticin-J homolog are two uncharacterised transport proteins and a multidrug resistant protein, thus presenting a method of export and immunity to the action of the protein.
The holin containing gene cluster presents a potential antibacterial mechanism of L. johnsonii KD1. This suggests that the protein is less likely to be directly associated with WSSV inhibition and may act indirectly by influencing the gut microbiome. On the other hand, the bacteriocin helveticin-J homolog proposes a potential direct WSSV inhibition mechanism, as bacteriocins with anti-viral properties have been observed.
One mechanism by which bacteriocins can exert their antiviral properties is via blocking host cell receptor sites and preventing viral entry64. The ability of the helveticin-J homolog to interfere with VP28 mediated cell entry was assessed via protein–protein docking. To be able to model these potential interactions, 3D structures of the proteins were needed. Structures of the monomer and trimer of VP28 (both determined by X-ray crystallography) were downloaded from Phyre2 and RCSB Protein Data Bank (RCSB PDB), respectively. Furthermore, the AlphaFold protein structure database contained an AlphaFold V2 generated model of a protein with 100% homology to the BV-BRC identified helveticin-J homolog (Lj_peg_533) named Q74KQ5. In addition, two models of the BAGEL4 helveticin-J homolog (Lj_6.3) were generated using the google collab AlphaFold2 software, one using a PDB70 model and one without a model (Lj_6.3.1 and Lj_6.3.2, respectively). Both models were similar with an RMSD of 0.003. In addition, the AlphaFold predicted structure of PmRab7 was used, as it has been demonstrated to interact with the monomer of VP2865.
Next, the LZerD protein docking webserver was used to generate docking models of each putative ligand (PmRab7, Lj_6.3.1, Lj_6.3.2 and Q74KQ5) with the monomer and trimer of VP28. As seen in Fig. 8, all putative ligands bind in the same region of both the VP28 monomer and trimer, thus suggesting that if these interactions were to occur then the helveticin-J homolog may be able to inhibit PmRab7 and VP28 interaction. This could therefore prevent WSSV infection, as PmRab7 binding with VP28 has been associated with WSSV infection of P. monodon65. Furthermore, P. vannamei has a Rab7 protein with 100% homology to PmRab7, so this interaction could be blocked in this organism too. In addition, as PmRab7 is associated with endosomal trafficking, it presents a potential different mechanism to direct inhibition of viral entry66. However, LZerD protein–protein docking works on the assumption that both proteins do interact. Therefore, molecular dynamics simulations should be carried out to assess the likelihood of each model of the helveticin-J homolog interacting with VP28.
Finally, this study suggests potential mechanisms behind the anti-WSSV properties demonstrated by L. johnsonii KD1. Thus, the list of metabolites identified in this study will be examined and tested for WSSV inhibition to identify the specific anti-WSSV mechanisms.
Conclusions
This study investigated the roles of the probiotic Lactobacillus johnsonii KD1 in WSSV inhibition. Firstly, shrimps treated with 100X L. johnsonii KD1 reduced shrimp cumulative mortality after WSSV infection through reduction of viral production and activation of shrimp immune systems. Secondly, the complete genome of this probiotic showed one AMR gene, not related to mobile genetic elements, and a type I restriction-modification system on the probiotic chromosome. This suggests a potential phage resistance mechanism and a limited chance of AMR dissemination into the environment through mobile genetic elements. Thirdly, CAZymes analysis identified genes coding for GT8 family proteins in L. johnsonii KD1 which could be associated with the biosynthesis of lipopolysaccharides with potential anti-WSSV properties. Interestingly, through protein–protein docking modelling it was seen that the homolog of the bacteriocin helveticin-J identified in L. johnsonii KD1 may be able to block viral entry. All the findings of this study suggest that the probiotic L. johnsonii KD1 is friendly to use in animal supplements and able to inhibit WSSV infection. Additionally, probiotic derived metabolites could be prepared as postbiotics for shrimp cultivation in the future.
Experimental procedures
Preparation of probiotic supplement for application in shrimp experiments
To prepare probiotic supplement, frozen stock of L. johnsonii KD1 in − 80 °C freezer was streaked on De Man–Rogosa–Sharpe (MRS) agar and incubated at 37 °C without shaking for 24 h. A single colony of L. johnsonii KD1 was inoculated into MRS broth and incubated overnight at 37 °C without shaking. A 1:100 dilution of L. johnsonii KD1 culture was transferred to a new medium and incubated at 37 °C without shaking for 2.5 h. Probiotic cells at 1X (4.5 × 108 CFU) and 100X (4.5 × 1010 CFU) were collected by centrifugation at 6000 rpm for 5 min and each probiotic pellet was resuspend in 1 mL medium prior to submersion into each shrimp tank.
Efficacy of the probiotic L. johnsonii KD1 against WSSV infection in shrimp
WSSV-infected tissues were prepared according to the previous study6. Specific pathogen free (SPF) white shrimp P. vannamei juveniles (20 g B.W.) provided by Charoen Pokphand Foods (CPF) Thailand were cultured in artificial seawater with salinity of 10 parts per thousand (ppt) with aeration and average water temperature of 28 ± 0.5 °C. The juveniles were acclimatized for three days before performing experiments. Shrimps were fed with WSSV-infected tissues at 10% of shrimp body weight. Then, shrimp muscles from moribund shrimps were collected and DNA was extracted to quantify WSSV copies/g tissues by qPCR. For WSSV challenge, SPF P. vannamei post larvae (0.03 g B.W.) provided by CPF were cultured in artificial seawater with salinity of 20 ppt with aeration and average water temperature of 28 ± 0.5 °C. The efficacy of the probiotic treatments was tested by directly adding the probiotic into shrimp tanks. Each shrimp tank was a glass tank sized 30 × 20 × 15 cm3 filled with 2 L seawater. Shrimps were divided into four groups with three tanks (n = 30 shrimps/tank/2 L seawater) containing (1) shrimps’ regimen with 1X L. johnsonii KD1 (4.5 × 108 CFU/2 L seawater), (2) 100X L. johnsonii KD1 (4.5 × 1010 CFU/2 L seawater), according to a previous study6, for five days before WSSV infection, (3) positive group meaning WSSV-treated shrimps without probiotic treatment, and (4) negative group meaning shrimps without any treatment (Supplementary Fig. 3A). During probiotic treatment for five days, water was not changed and then 100% water change before WSSV challenge. WSSV-infected tissues were provided to the shrimp at approximately log WSSV copies of 10.2–10.7/1 g tissues/replicate according to the previous study6. Numbers of live shrimps were recorded daily until 100% of mortality in the positive group. Percentage shrimp cumulative mortality was calculated from remaining percentage of cumulative shrimp survival after oral challenge with WSSV-infected tissue. Additionally, viral loads of WSSV were quantitated from three survived shrimps per group at 3-day post infection (dpi.). During experiment, the shrimps were fed with normal feed twice daily at the amount of 10% shrimp body weight per day and 100% water changes were performed every 2 days. For water quality, sufficient aeration, average water temperature of 28 ± 0.5 °C, water color, odor, pH and alkalinity were checked every day. Additionally, shrimp feces were removed every day to control water quality.
Detection and quantitation of WSSV in shrimps by qPCR
To quantitate WSSV in shrimps, shrimp DNA was collected and extracted according to the previous study6. One hundred ng of shrimp DNA was used for qPCR with reactions consisting of 1X QuantiNova™ SYBR® Green PCR Master Mix (Qiagen), 0.7 µM WSSV229_F, 0.7 µM WSSV447_R primers (Supplementary Table 11) and adjusted with nuclease-free water into 20 µL final volume. Then, Rotor-Gene® Q PCR machine (Qiagen) and Rotor-Gene Q 5plex HRM Platform software were used to run qPCR and analyze the results. The viral copies were calculated from a standard curve of WSSV specific 448 amplicons at 109 to 10 copies. If WSSV copies were lower than 10 copies, they were considered to be non-detectable (ND) due to limit of detection (LOD).
Expression analysis of shrimp immune genes for probiotic treatments
To assess shrimp immune activation by probiotics we performed experiments according to the previous study6,67. Briefly, white shrimp P. vannamei post larvae (average size of 0.03 g) were treated with L. johnsonii KD1 at 100X dose (4.5 × 1010 CFU/2 L seawater) to investigate expression of shrimp innate immune genes, namely genes coding for peroxinectin (PX), serine protease (SP), prophenoloxidase-1 (proPO I), and anti-lipopolysaccharide factor-1 (ALF-1). Post-larvae shrimps were divided into two groups receiving 100X L. johnsonii KD1, and no bacteria (negative control). Each group contained two replicates, n = 30 shrimps/replicate/2 L seawater. Shrimps were treated with the probiotic for five days except the negative group and water was not changed during probiotic treatments. At day 6 (or day 1 post probiotic treatment) and day 8 (or day 3 post probiotics treatment), four shrimps per group were collected (Supplementary Fig. 3B). Shrimp RNA was extracted and qRT-PCR was performed according to the previous study6. The lists of primers are shown in Supplementary Table 11. Absolute copies of each gene were calculated based on a standard curve of 109 to 10 copies of plasmids containing the shrimp immune genes. Then, each gene was normalized by a housekeeping gene coding for β-actin RNA. Finally, fold changes of shrimp gene expression were quantitated from expression ratios of shrimps treated with probiotic treatments against shrimps without probiotic treatments (negative control).
DNA extraction of L. johnsonii KD1
L. johnsonii KD1 was cultured in 15 ml of MRS broth at 37 °C without shaking for 24 h. After that, 1.5 × 109 CFU/mL was utilized for genomic DNA extraction using Exgene™ Cell SV according to GeneAll® Exgene™ manual instruction. Genomic DNA was quality checked by 0.8% gel electrophoresis and DNA concentration (ng/μL) was measured using UV absorbance at OD260/ OD280 and OD260/230 with a NanoDrop™ spectrophotometer (Thermo Scientific) and Qubit dsDNA DNA BRAssay kit (Life Technologies) followed by storage at − 20 °C until use.
Library preparation and sequencing
One µg of genomic DNA was used to prepare a DNA library for long read sequencing by using the ligation sequencing gDNA kit (SQK-LSK110, Oxford nanopore technologies) according to the guidelines. Briefly, one µg of genomic DNA in 47 µL was added into a reaction consisting of 1 µL DNA CS, 3.5 µL NEBNext FFPE DNA Repair Buffer, 2 µL NEBNext FFPE DNA Repair Mix, 3.5 µL Ultra II End-prep Reaction Buffer, and 3 µL Ultra II End-prep Enzyme Mix followed by incubation at 20 °C for 5 min and 65 °C for 5 min. To clean up genomic DNA, 60 µL of AMPure XP beads was added and incubated for 5 min at room temperature followed by a wash with 70% ethanol. DNA pellet was dissolved with 61 µL nuclease-free water. To ligate the adapter, 60 µL DNA was mixed with 25 µL Ligation Buffer (LNB), 10 µL NEBNext Quick T4 DNA Ligase, and 5 µL Adapter Mix F (AMX-F) followed by incubation at room temperature for 10 min. Genomic DNA was cleaned up again with 40 µL of AMPure XP beads. 250 μL Long Fragment Buffer (LFB) was added. Then, DNA libraries at 5–50 fmol were loaded and sequenced by MinION and MinKNOW software v3.5.40 (Oxford Nanopore Technologies).
For short reads sequencing, 100 ng of genomic DNA was used to prepare DNA libraries for Illumina sequencing by using TruSeq DNA Nano kit (Illumina) according to TruSeq® Nano DNA Library Preparation guidelines. Briefly, 100 ng of genomic DNA was fragmented with Covaris settings. Then, DNA fragments were cleaned up with adding 80 μL SPB, 200 μL of 80% ethanol, and 62.5 μL RSB, respectively. DNA fragments was treated to blunt ends using 40 μL End Repair Mix 2 and incubated at 30 °C for 30 min followed by an incubation on ice. DNA fragments were size selected using different ratios of the SPB and performed adenylate 3ʹ ends by adding 12.5 μL ATL. To ligate adapters, 2.5 μL of each RSB, LIG2, and DNA adapters was added and cleaned up with SPB, 80% ethanol and RSB, respectively. Then, DNA fragments were enriched by adding 20 μL EPM. Finally, DNA libraries were normalized and pooled. Paired-end sequencing was performed on Illumina HiSeq platform.
De novo assembly and annotation
Raw reads from Illumina and Nanopore were presented in Supplementary Table 12. Long reads from nanopore sequencing were concatenated by concatenate datasets tail-to-head (cat) version 0.1.1 and adaptors were removed by Porechop version 0.2.4. Long reads with a length of less than 1000 bp were removed by filtlong version 0.2.1. For short reads from Illumina sequencing, Trim Galore version 0.6.7 was used to remove adaptors from raw short reads. Then, long and short reads were combined for hybrid de novo assembly by Unicycler version 0.4.8 pipeline68. Complete genomes were annotated by the Rapid Annotation using Subsystem Technology toolkit (RASTtk) in the Bacterial and Viral Bioinformatics Resource Center (BV-BRC) version 3.25.369 and visualized using CG View (Circular Genome Viewer) in Proksee44.
Species identification and phylogenetic analysis
The chromosomal sequence was used for species determination by Ribosomal Multilocus Sequence Typing (rMLST)70 and for molecular typing and microbial genome diversity by PubMLST71. Plasmid sequence was characterized using BLASTn in National Center for Biotechnology Information (NCBI). Then, a phylogenetic tree was constructed based on 500 single-copy genes from 21 L. johnsonii genomes consisting of L. johnsonii KD1 (This study), L. johnsonii N6.2 (CP006811), L. johnsonii BIO5467 (WBNZ00000000), L. johnsonii Byun-jo-01 (CP029614), L. johnsonii FI9785 (FN298497), L. johnsonii G2A (CP040854), L. johnsonii DC22.2 (CP039261), L. johnsonii CNCM I-4884 (JAIQXC000000000), L. johnsonii NCK2677 (CP059055), L. johnsonii MT4 (JAJQJG000000000), L. johnsonii pf01 (AFQJ00000000), L. johnsonii ATCC 33200 (ACGR00000000), L. johnsonii GHZ10a (CP062068), L. johnsonii DPC 6026 (CP002464), L. johnsonii IDCC9203 (CP031701), L. johnsonii ZLJ010 (CP032680), L. johnsonii NCC 533 (AE017198), L. johnsonii UMNLJ21(CP021703), L. johnsonii UMNLJ22 (CP021704), L. johnsonii BS15 (CP016400), and L. johnsonii W1 (LSNG00000000) using Randomized Axelerated Maximum Likelihood (RAxML) in the Phylogenetic Tree Service of BV-BRC version 3.25.369.
Prediction of antimicrobial resistance, mobile genetic elements and clustered regularly interspaced short palindromic repeats (CRISPR)-Cas system, pathogenicity
Both the chromosome and plasmid sequences were investigated for antimicrobial resistance genes by ResFinder-4.172,73,74 and CARD (Comprehensive Antibiotic Resistance Database)75. Additionally, prophages, insertion sequences, and transposons were identified by PHASTER (PHAge Search Tool Enhanced Release)76,77 and MGEfinder version 1.0.378, respectively. Finally, CRISPR-Cas system was detected by using CRISPRCasFinder tool version 4.2.3079 and pathogenicity was predicted using PathogenFinder version 1.180.
Comparative genomics
The list of 21 L. johnsonii genomes as previously described were analyzed by comparative genomics using the protein family sorter program with PATRIC genus-specific families (PLfams) in BV-BRC version 3.25.369 to identify core and unique coding sequences (CDS) in L. johnsonii KD1. Beyond comparative genomics of L. johnsonii strains, genomes of L. johnsonii KD1 and Lactiplantibacillus plantarum, previously named Lactobacillus plantarum81, were compared to identify unique genes of L. johnsonii KD1 by using the protein family sorter program with PATRIC cross-genus families (PGfams) and MicrobeAnnotator V 2.0.5 (utilising Kofam and Swiss-Prot databases)82. Additionally, the dbCAN2 web server37 was used to annotate the carbohydrate-active enzymes (CAZymes) in the L. johnsonii KD1 and two L. plantarum ATCC 14917 genomes (1 strain isolated from pickled cabbage (AZEJ00000000.1, Shanghai Majorbio) and a strain isolated from a human gut (ACGZ00000000.2, Baylor College of Medicine) using all three gene annotation tools (HMMER: dbCAN, DIAMOND: CAZy and HMMER: dbCAN-sub) plus the CGC finder tool. Annotations produced by two or more gene annotation tools were accepted. Heatmaps were then generated to compare the CAZyme profiles of each genome.
Cellular adhesion protein identification
Lactobacillus cellular adhesion proteins were identified in the literature47,48 and sequences were obtained from NCBI83. The presence of cell adhesion proteins was identified in L. johnsonii KD1 and the two L. plantarum ATCC 14917 isolates using the BV-BRC BLASTp tool. Heatmaps were then generated using the bit-score of the tops hits.
Prediction of antimicrobial peptide production
The genomes of L. johnsonii KD1 and L. plantarum ATCC 14917 were analyzed for the presence of genes encoding putative antimicrobials by BAGEL484 and AntiSMASH version 6.1.185. Further analysis of the genes identified surrounding the gassericin T cluster homolog was conducted using NCBI BLASTp, UniProt BLASTp and compared to the BV-BRC L. johnsonii KD1 annotation86,87.
Bacteriocin helveticin-J homolog: VP28 interaction modelling:
Structures of the VP28 monomer and trimer (d2ed6a1 and 2ED6, respectively) were collected from Phyre288 and RCSB Protein Data Bank (RCSB PDB). The structures of PmRab7 and a homolog of the bacteriocin helveticin-J from L. johnsonii (strain CNCM I-12250/La1/NCC 533) (Q74KQ5) (which has 100% sequence similarity to the BV-BRC predicted sequence of the helveticin-J homolog from L. johnsonii KD1) were obtained from the AlphaFold protein structure database89. Furthermore, two model structures of the BAGEL4 predicted sequence of the helveticin-J homolog in L. johnsonii KD1 were produced using the google collab AlphaFold V2, with a PDB70 template or no template filter selected. Structures were named Lj_6.3.1 and Lj_6.3.2, respectively. The LZerD web server90 was utilised to produce models of PmRab7, Q74KQ5, Lj_6.3.1 and Lj_6.3.2 interactions with the monomer and trimer of VP28. The 3D structures of respective proteins were used as inputs with the clustering cutoff (Å) set to 4 and surface reduction cutoff set to 1e-4 (default settings).
Statistical analysis
Student’s T-test, Mann–Whitney Test, one-way ANOVA on SPSS statistics with Duncan post-hoc test were used in data analyses. Statistically significance is reported when p-value is less than 0.05.
Ethics statement
All applicable institution guideline MUSC-IACUC, protocol no. MUSC63-003-511 and MUSC66-023-653 for the care and use of animals were followed.
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information file). The complete genome and plasmid sequence data have been submitted to the GenBank databases under BioProject PRJNA936661 and accession numbers CP118625 and CP118626, respectively.
References
Asche, F. et al. The economics of shrimp disease. J. Invertebr. Pathol. 186, 107397 (2021).
El-Saadony, M. T. et al. Shrimp production, the most important diseases that threaten it, and the role of probiotics in confronting these diseases: A review. Res. Vet. Sci. 144, 126–140 (2022).
Shinn, A. et al. Asian shrimp production and the economic costs of disease. Asian Fish. Sci. 31, 29–58 (2018).
Chattaraj, S., Ganguly, A., Mandal, A. & Das Mohapatra, P. K. A review of the role of probiotics for the control of viral diseases in aquaculture. Aquacult. Int. 30, 2513–2539 (2022).
Patil, P. K. et al. Economic loss due to diseases in Indian shrimp farming with special reference to Enterocytozoon hepatopenaei (EHP) and white spot syndrome virus (WSSV). Aquactic 533, 736231 (2021).
Dekham, K. et al. Probiotics expressing double-stranded RNA targeting VP28 efficiently protect shrimps from WSSV infection. Aquac. Rep. 23, 101067 (2022).
George, F. et al. Occurrence and dynamism of lactic acid bacteria in distinct ecological niches: A multifaceted functional health perspective. Front. Microbiol. 9, 2899 (2018).
Ringo, E. et al. Lactic acid bacteria in finfish: An update. Front. Microbiol. 2018, 9 (1818).
Lubeck, M. & Lubeck, P. S. Application of lactic acid bacteria in green biorefineries. FEMS Microbiol. Lett. 366, 024 (2019).
Vazquez-Munoz, R. et al. Insights from the Lactobacillus johnsonii genome suggest the production of metabolites with antibiofilm activity against the pathobiont Candida albicans. Front. Microbiol. 13, 853762 (2022).
Zheng, D. et al. Immune responses in pregnant sows induced by recombinant Lactobacillus johnsonii expressing the COE protein of porcine epidemic diarrhea virus provide protection for piglets against PEDV infection. Viruses 14, 7 (2021).
Zou, Y. J. et al. Lactobacillus johnsonii L531 ameliorates Escherichia coli-induced cell damage via inhibiting NLRP3 inflammasome activity and promoting ATG5/ATG16L1-mediated autophagy in porcine mammary epithelial Cells. Vet. Sci. 7, 112 (2020).
Wang, W. et al. Screening of lactic acid bacteria with inhibitory activity against ETEC K88 as feed additive and the effects on sows and piglets. Animals 11, 1719 (2021).
Olnood, C. G., Beski, S. S. M., Choct, M. & Iji, P. A. Use of Lactobacillus johnsonii in broilers challenged with Salmonella sofia. Anim. Nutr. 1, 203–212 (2015).
Manes-Lazaro, R. et al. Administration of Lactobacillus johnsonii FI9785 to chickens affects colonisation by Campylobacter jejuni and the intestinal microbiota. Br. Poult. Sci. 58, 373–381 (2017).
Richards, P. J. et al. Galacto-oligosaccharides modulate the juvenile gut microbiome and innate immunity to improve broiler chicken performance. MSystems 5, e00827 (2020).
Torrecillas, S. et al. Effect of fishmeal and fish oil replacement by vegetable meals and oils on gut health of European sea bass (Dicentrarchus labrax). Aquaculture 468, 386–398 (2017).
Leyva-Madrigal, K. Y., Luna-González, A., Escobedo-Bonilla, C. M., Fierro-Coronado, J. A. & Maldonado-Mendoza, I. E. Screening for potential probiotic bacteria to reduce prevalence of WSSV and IHHNV in whiteleg shrimp (Litopenaeus vannamei) under experimental conditions. Aquaculture 322, 16–22 (2011).
Cerenius, L. & Soderhall, K. The prophenoloxidase-activating system in invertebrates. Immunol. Rev. 198, 116–126 (2004).
Chiu, C.-H., Guu, Y.-K., Liu, C.-H., Pan, T.-M. & Cheng, W. Immune responses and gene expression in white shrimp, Litopenaeus vannamei, induced by Lactobacillus plantarum. Fish Shellfish Immunol. 23, 364–377 (2007).
Tassanakajon, A. et al. Shrimp humoral responses against pathogens: antimicrobial peptides and melanization. Dev. Comp. Immunol. 80, 81–93 (2018).
Maeda, M. et al. Isolation of lactic acid bacteria from kuruma shrimp (Marsupenaeus japonicus) intestine and assessment of immunomodulatory role of a selected strain as probiotic. Mar. Biotechnol. 16, 181–192 (2014).
Li, H. et al. RNAi screening identifies a new Toll from shrimp Litopenaeus vannamei that restricts WSSV infection through activating Dorsal to induce antimicrobial peptides. PLoS Pathog. 14, e1007109 (2018).
Zhang, W. et al. Complete genome sequencing and comparative genome characterization of Lactobacillus johnsonii ZLJ010, a potential probiotic with health-promoting properties. Front. Genet. 10, 812 (2019).
Moradi, J. et al. Characterization of the resistome in Lactobacillus genomic sequences from the human gut. J. Glob. Antimicrob. Resist. 30, 451–458 (2022).
Výrostková, J. et al. Antimicrobial resistance of Lactobacillus johnsonii and Lactobacillus zeae in raw milk. Processes 8, 1627 (2020).
Dec, M., Nowaczek, A., Stepien-Pysniak, D., Wawrzykowski, J. & Urban-Chmiel, R. Identification and antibiotic susceptibility of lactobacilli isolated from turkeys. BMC Microbiol. 18, 168 (2018).
Dec, M., Urban-Chmiel, R., Stepien-Pysniak, D. & Wernicki, A. Assessment of antibiotic susceptibility in Lactobacillus isolates from chickens. Gut Pathog. 9, 54 (2017).
Chen, Z. et al. A type I restriction modification system influences genomic evolution driven by horizontal gene transfer in Paenibacillus polymyxa. Front. Microbiol. 12, 709571 (2021).
Seib, K. L. & Jennings, M. P. Chapter 24: Epigenetics of infectious diseases. In Medical epigenetics (ed. Tollefsbol, T. O.) 443–458 (Academic Press, 2016).
Vasu, K. & Nagaraja, V. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 77, 53–72 (2013).
Guinane, C. M. et al. Host specific diversity in Lactobacillus johnsonii as evidenced by a major chromosomal inversion and phage resistance mechanisms. PLoS ONE 6, e18740 (2011).
Garneau, J. E. & Moineau, S. Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb. Cell Fact. 10(Suppl 1), S20 (2011).
Huang, T. J., Liu, S. H., Kuo, Y. C., Chen, C. W. & Chou, S. C. Antiviral activity of chemical compound isolated from Artemisia morrisonensis against hepatitis B virus in vitro. Antiviral Res. 101, 97–104 (2014).
Li, C., Yang, M. C., Hong, P. P., Zhao, X. F. & Wang, J. X. Metabolomic profiles in the intestine of shrimp infected by white spot syndrome virus and antiviral function of the metabolite linoleic acid in shrimp. J. Immunol. 206, 2075–2087 (2021).
Ryang, J., Yan, Y., Song, Y., Liu, F. & Ng, T. B. Anti-HIV, antitumor and immunomodulatory activities of paclitaxel from fermentation broth using molecular imprinting technique. AMB Express 9, 194 (2019).
Zhang, H. et al. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101 (2018).
Ausland, C. et al. dbCAN-PUL: A database of experimentally characterized CAZyme gene clusters and their substrates. Nucleic Acids Res. 49, D523–D528 (2021).
Lee, I. C. et al. Strain-specific features of extracellular polysaccharides and their impact on Lactobacillus plantarum-host interactions. Appl. Environ. Microbiol. 82, 3959–3970 (2016).
Ruiz, L., Margolles, A. & Sanchez, B. Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front. Microbiol. 4, 396 (2013).
Yao, W., Yang, L., Shao, Z., Xie, L. & Chen, L. Identification of salt tolerance-related genes of Lactobacillus plantarum D31 and T9 strains by genomic analysis. Ann. Microbiol. 70, 1–14 (2020).
O’Flaherty, S., Briner Crawley, A., Theriot, C. M. & Barrangou, R. The Lactobacillus bile salt hydrolase repertoire reveals niche-specific adaptation. MSphere 3, e00140 (2018).
McAuliffe, O., Cano, R. J. & Klaenhammer, T. R. Genetic analysis of two bile salt hydrolase activities in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71, 4925–4929 (2005).
Boucard, A. S., Florent, I., Polack, B., Langella, P. & Bermudez-Humaran, L. G. Genome sequence and assessment of safety and potential probiotic traits of Lactobacillus johnsonii CNCM I-4884. Microorganisms 10, 273 (2022).
Papadimitriou, K. et al. Discovering probiotic microorganisms: In vitro, in vivo, genetic and omics approaches. Front. Microbiol. 6, 58 (2015).
Wang, Y., Moon, A., Huang, J., Sun, Y. & Qiu, H. J. Antiviral effects and underlying mechanisms of probiotics as promising antivirals. Front. Cell Infect. Microbiol. 12, 928050 (2022).
Surve, S., Shinde, D. B. & Kulkarni, R. Isolation, characterization and comparative genomics of potentially probiotic Lactiplantibacillus plantarum strains from Indian foods. Sci. Rep. 2022, 12 (1940).
Muscariello, L., De Siena, B. & Marasco, R. Lactobacillus cell surface proteins involved in interaction with mucus and extracellular matrix components. Curr. Microbiol. 77, 3831–3841 (2020).
Moon, A. et al. Lactic acid bacteria as mucosal immunity enhancers and antivirals through oral delivery. Appl. Microbiol. 2, 837–854 (2022).
Nahui Palomino, R. A., Zicari, S., Vanpouille, C., Vitali, B. & Margolis, L. Vaginal Lactobacillus inhibits HIV-1 replication in human tissues ex vivo. Front. Microbiol. 8, 906 (2017).
Al Kassaa, I., Hober, D., Hamze, M., Chihib, N. E. & Drider, D. Antiviral potential of lactic acid bacteria and their bacteriocins. Probiotics Antimicrob. Proteins 6, 177–185 (2014).
Tiwari, S. K. et al. Probiotics at war against viruses: What is missing from the picture?. Front. Microbiol. 2020, 11 (1877).
Conti, C., Malacrino, C. & Mastromarino, P. Inhibition of herpes simplex virus type 2 by vaginal lactobacilli. J. Physiol. Pharmacol. 60(Suppl 6), 19–26 (2009).
Biliavska, L., Pankivska, Y., Povnitsa, O. & Zagorodnya, S. Antiviral activity of exopolysaccharides produced by lactic acid bacteria of the genera Pediococcus, Leuconostoc and Lactobacillus against human adenovirus type 5. Medicina 55, 519 (2019).
Werning, M. L. et al. Biological functions of exopolysaccharides from lactic acid bacteria and their potential benefits for humans and farmed animals. Foods 11, 1284 (2022).
Deo, D., Davray, D. & Kulkarni, R. A diverse repertoire of exopolysaccharide biosynthesis gene clusters in Lactobacillus revealed by comparative analysis in 106 sequenced genomes. Microorganisms 7, 444 (2019).
Horn, N. et al. Spontaneous mutation reveals influence of exopolysaccharide on Lactobacillus johnsonii surface characteristics. PLoS ONE 8, e59957 (2013).
Luna-González, A. et al. The prebiotic inulin increases the phenoloxidase activity and reduces the prevalence of WSSV in whiteleg shrimp (Litopenaeus vannamei) cultured under laboratory conditions. Aquaculture 362, 28–32 (2012).
Zhou, L. et al. Dietary prebiotic inulin benefits on growth performance, antioxidant capacity, immune response and intestinal microbiota in Pacific white shrimp (Litopenaeus vannamei) at low salinity. Aquaculture 518, 734847 (2020).
Anwar, M. A., Kralj, S., van der Maarel, M. J. & Dijkhuizen, L. The probiotic Lactobacillus johnsonii NCC 533 produces high-molecular-mass inulin from sucrose by using an inulosucrase enzyme. Appl. Environ. Microbiol. 74, 3426–3433 (2008).
Anwar, M. A. et al. The role of conserved inulosucrase residues in the reaction and product specificity of Lactobacillus reuteri inulosucrase. FEBS J. 279, 3612–3621 (2012).
Lakshmi, B., Viswanath, B. & Sai Gopal, D. V. Probiotics as antiviral agents in shrimp aquaculture. J. Pathog. 2013, 424123 (2013).
Saier, M. H. Jr. & Reddy, B. L. Holins in bacteria, eukaryotes, and archaea: Multifunctional xenologues with potential biotechnological and biomedical applications. J. Bacteriol. 197, 7–17 (2015).
Umair, M. et al. Probiotic-based bacteriocin: Immunity supplementation against viruses an updated review. Front. Microbiol. 13, 876058 (2022).
Sritunyalucksana, K., Wannapapho, W., Lo, C. F. & Flegel, T. W. PmRab7 is a VP28-binding protein involved in white spot syndrome virus infection in shrimp. J. Virol. 80, 10734–10742 (2006).
Ongvarrasopone, C., Chanasakulniyom, M., Sritunyalucksana, K. & Panyim, S. Suppression of PmRab7 by dsRNA inhibits WSSV or YHV infection in shrimp. Mar. Biotechnol. 10, 374–381 (2008).
Won, S. et al. Evaluation of potential probiotics Bacillus subtilis WB60, Pediococcus pentosaceus, and Lactococcus lactis on growth performance, immune response, gut histology and immune-related genes in whiteleg shrimp, Litopenaeus vannamei. Microorganisms 8, 281 (2020).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017).
Olson, R. D. et al. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 51, D678–D689 (2023).
Jolley, K. A. et al. Ribosomal multilocus sequence typing: Universal characterization of bacteria from domain to strain. Microbiology 158, 1005–1015 (2012).
Jolley, K. A., Bray, J. E. & Maiden, M. C. J. Open-access bacterial population genomics: BIGSdb software, the PubMLSTorg website and their applications. Wellcome Open Res. 3, 124 (2018).
Bortolaia, V. et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500 (2020).
Camacho, C. et al. BLAST+: Architecture and applications. BMC Bioinform. 10, 421 (2009).
Zankari, E. et al. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768 (2017).
Alcock, B. P. et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525 (2020).
Arndt, D. et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16-21 (2016).
Zhou, Y., Liang, Y., Lynch, K. H., Dennis, J. J. & Wishart, D. S. PHAST: A fast phage search tool. Nucleic Acids Res. 39, W347-352 (2011).
Johansson, M. H. K. et al. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 76, 101–109 (2021).
Grissa, I., Vergnaud, G. & Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52-57 (2007).
Cosentino, S., Voldby Larsen, M., Moller Aarestrup, F. & Lund, O. PathogenFinder–distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE 8, e77302 (2013).
Zheng, J. et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858 (2020).
Ruiz-Perez, C. A., Conrad, R. E. & Konstantinidis, K. T. MicrobeAnnotator: A user-friendly, comprehensive functional annotation pipeline for microbial genomes. BMC Bioinform. 22, 11 (2021).
Sayers, E. W. et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 50, D20–D26 (2022).
van Heel, A. J. et al. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 46, W278–W281 (2018).
Blin, K. et al. antiSMASH 60: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35 (2021).
Zhao, X. & Kuipers, O. P. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genomics 17, 882 (2016).
UniProt, C. UniProt: The universal protein knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531 (2023).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Varadi, M. et al. AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
Christoffer, C., Bharadwaj, V., Luu, R. & Kihara, D. LZerD protein–protein docking webserver enhanced with de novo structure prediction. Front. Mol. Biosci. 8, 724947 (2021).
Acknowledgements
Authors would like to thank the Thailand Toray Science Foundation, the Science Achievement Scholarship of Thailand, the Faculty of Science (Mahidol University), and the Center of Excellence for Shrimp Molecular Biology and Biotechnology (Centex Shrimp). Furthermore, we are also grateful to the South Coast Biosciences Doctoral Training Partnership (SoCoBio DTP) funded by Biotechnology and Biological Sciences Research Council (BBSRC) to SMJ.
Funding
This research project was funded by Thailand Toray Science Foundation (TTSF Year 2021) to SC, Science Achievement Scholarship of Thailand (SAST) to KD, and South Coast Biosciences Doctoral Training Partnership (SoCoBio DTP) to SMJ.
Author information
Authors and Affiliations
Contributions
K.D.: Methodology, Data analyses, Writing; S.M.J.: Methodology, Data analyses, Writing; S.J.: Methodology; P.C.: Methodology; N.T.: Methodology; R.I.: Methodology; P.D.: Providing bacterial strains and suggestions; S.S.: Providing supervision, Writing; V.S.: Conceptualizing the study, Providing supervision, Writing; S.C.: Conceptualizing the study, Providing supervision, Writing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dekham, K., Jones, S.M., Jitrakorn, S. et al. Functional and genomic characterization of a novel probiotic Lactobacillus johnsonii KD1 against shrimp WSSV infection. Sci Rep 13, 21610 (2023). https://doi.org/10.1038/s41598-023-47897-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-023-47897-w