Abstract
Manipulative behaviour that consists of touching or close contact with ears or tails of pen mates is common in pigs and can become damaging. Manipulative behaviour was analysed from video recordings of 45-day-old pigs, and 15 manipulator-control pairs (n = 30) were formed. Controls neither received nor performed manipulative behaviour. Rectal faecal samples of manipulators and controls were compared. 16S PCR was used to identify Lactobacillaceae species and 16S amplicon sequencing to determine faecal microbiota composition. Seven culturable Lactobacillaceae species were identified in control pigs and four in manipulator pigs. Manipulators (p = 0.02) and females (p = 0.005) expressed higher Lactobacillus amylovorus, and a significant interaction was seen (sex * status: p = 0.005) with this sex difference being more marked in controls. Females (p = 0.08) and manipulator pigs (p = 0.07) tended to express higher total Lactobacillaceae. A tendency for an interaction was seen in Limosilactobacillus reuteri (sex * status: p = 0.09). Results suggest a link between observed low diversity in Lactobacillaceae and the development of manipulative behaviour.
Similar content being viewed by others
Introduction
Different forms of damaging biting behaviour are common in pigs1, and can for instance be seen in psychologically stressed animals2. Biting behaviour is typically directed to tails and ears1,3,4. Four types of tail biting have been suggested: (1) obsessive, (2) sudden and forceful, (3) two-stage5, and (4) epidemic tail biting6. Tail biting decreases the welfare of animals7 and affects the farmer’s income8. The precise mechanism behind tail biting remains poorly understood, but research suggests that it is a multifactorial problem8. Chronic stress, salt deficiency, nutritional deprivation3, poor access to feed7, diet formulation, health problems, environmental discomfort, high stocking density, and inadequate rooting material8 are among the known risk factors for tail biting. Pig-directed manipulative behaviour can be defined as manipulation of tails/ears without always resulting in visible wounds9. For example, tail-in-mouth behaviour is linked to damaging behaviour and often precedes tail biting10. Manipulative and damaging behaviour are thus probably behaviours caused by similar motivational factors but expressed in different severity ranging from mild occasional manipulation to outbreaks with severely damaged ears and tails8. Current research suggests a relationship between negative behavioural expression and the gut microbiota. Van der Eijk and colleagues11, for instance, showed feather-pecking hens to have a lower abundance of Lactobacillus than their non-feather-pecking peers. Brunberg and colleagues12 suggested a link between tail biting in pigs and the gut microbiota composition, which was supported by recent studies that found structural13 and compositional13,14 differences in microbiota between tail biters and control groups.
Lactobacillaceae play an important role in modifying the host epigenome and in producing beneficial metabolites such as short-chain fatty acids and vitamins15. Microbial development is rapid in early life16, requiring adequate environmental exposure for modulating the host immunity17. Vertical transmission of microbes is known to begin in some species, e.g. cattle, equine, and human, during pregnancy18,19,20,21, and to continue during lactation22,23,24. Colostrum is a key contributor to faecal Lactobacillaceae composition, influencing it long-term25,26. Besides diet, medication has a long-term impact on alpha diversity (Shannon diversity and observed species22 and Faith’s phylogenetic diversity27) and can lead to its depletion28. A decreased diversity may increase susceptibility to infections, e.g. due to overgrowth of opportunistic microbes in the gut28.
As microbial metabolites play a central role in the interaction between microbes and their hosts29, the gut microbiota influences the brain via the microbiota–gut–brain axis30 by the vagus nerve, circulatory system, and immune system30,31. Interestingly, microbial metabolites have been shown to influence depression in humans through multiple pathways32. The microbiota produces30,33,34 and regulates31 neurotransmitters such as serotonin35. Varying serotonin concentrations have been associated with anxiety and aggressiveness in different species36,37. In pigs, low concentrations have been associated with pessimistic-like behaviour38, and lower whole blood and platelet serotonin levels have been associated with tail biters compared to neutral, victim or biter/victim pigs39. Further, Valros and colleagues40 found evidence of a tendency for increased serotonin metabolism in tail-biting pigs, while other studies failed to support this41. Extensive research is required to help us to better understand the interaction between behaviour and intestinal microbiota and to find ways to alleviate tail biting in pigs.
Our aim was to identify pigs that perform tail and/or ear manipulation (manipulator pigs) and to compare their faecal microbiota with that of control pigs not manifesting such behaviour. We expected differences in the faecal microbiota between these two behavioural phenotypes. Specifically, we hypothesized finding less culturable Lactobacillaceae species in manipulator pigs. Furthermore, we hypothesized observing decreased total alpha diversity (based on Shannon and Chao1 indices and observed species richness) of microbiota in manipulator pigs compared with control pigs.
Results
Observed behaviour of pigs
Manipulator pigs (n = 15) performed on average two (range 1–3) different manipulative behaviours during the total of 66 min observations per pig (Supplementary Table S1), mainly directed towards ears. While all manipulator pigs manipulated ears, only two of them bit the tails of their pen mates (Supplementary Table S1). Five manipulator pigs received on average one (range 1–2) type of manipulative behaviour during the 66-min observation time. All performed and received manipulative behaviours of the study pigs are listed in Supplementary Table S1. None of the control pigs performed or received manipulative behaviour during the observation time.
Faecal Lactobacillaceae composition
The following seven culturable Lactobacillaceae species were identified in control pigs (in descending order of prevalence): Lactobacillus amylovorus, Limosilactobacillus reuteri, Lactobacillus johnsonii, Limosilactobacillus mucosae, Lactobacillus delbrueckii, Limosilactobacillus pontis, and Ligilactobacillus salivarus. Four species were identified in both manipulator and control pigs (in descending order of prevalence): L. amylovorus, L. reuteri, L. johnsonii, and L. mucosae. Control pigs tended (t28 = 1.7, p = 0.099) to have a higher average number of identified Lactobacillaceae species (1.8 species, SD 0.78) than manipulators (1.3 species, SD 0.72).
In total, 50% of isolated Lactobacillaceae remained unidentified in manipulator pigs and 54% in control pigs. Overall, manipulator pigs had a higher number of total Lactobacillaceae species (mean 2.3 × 109 colony-forming units (CFU)/ml; range 2 × 108–5.6 × 109) than control pigs (mean 1.2 × 109 CFU/ml; range 1.4 × 108–3.8 × 109) (Fig. 1). L. amylovorus was identified in 87% and L. reuteri in 43% of the samples regardless of the behaviour of the pig. Other Lactobacillaceae species were identified in 3–10% of samples, with the control group expressing higher numbers of these Lactobacillaceae (Fig. 1). Based on the Mann–Whitney U-test, no significant differences (p > 0.1) were found in abundance of L. johnsonii, L. mucosae, L. delbrueckii, L. pontis, or L. salivarius between manipulator and control pigs (Fig. 1).
According to the linear mixed models, both female (F1,26 = 3.3, p = 0.08, female 1.98 × 106 CFU/ml (SE 5.01 × 105) vs. barrow 1.22 × 106 CFU/ml (SE 4.49 × 105)) and manipulator pigs (F1,26 = 3.6, p = 0.07, manipulator 1.99 × 106 CFU/ml (SE 4.68 × 105) vs. control 1.21 × 106 CFU/ml (SE 4.8 × 105)) tended to have higher colony counts of total Lactobacillaceae species.
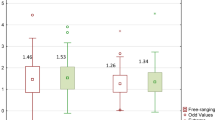
Female (F1,18 = 10.0, p = 0.005) and manipulator pigs (F1,17 = 6.5, p = 0.02) expressed higher colony counts of L. amylovorus than barrows and control pigs. In addition, a significant interaction sex * status (F1,18 = 9.9, p = 0.005) was seen (Fig. 2). The linear mixed model for L. reuteri showed a tendency for an interaction sex * status (F1,9 = 3.6, p = 0.09) (Fig. 3).
Amount of faecal Lactobacillus amylovorus in manipulator and control pigs. Back-transformed EM means of L. amylovorus (with 95% confidence interval) in manipulator (n = 15) and control pigs (n = 15) of both sexes (Linear mixed model, interaction sex * status: F1,18 = 9.9, p = 0.005). Middle line shows median, x shows the mean, dots show outliers, and whiskers show minimum and maximum values.
Amount of faecal Limosilactobacillus reuteri in manipulator and control pigs. Back-transformed EM means of L. reuteri (with 95% confidence interval) in manipulator (n = 15) and control pigs (n = 15) of both sexes (Linear mixed model, interaction sex * status: F1,9 = 3.6, p = 0.09). Middle line shows median, x shows the mean, dots show outliers, and whiskers show minimum and maximum values.
Microbiota analysis
No statistical differences in alpha diversity were observed between manipulator and control pigs (Shannon index: t26 = 1.22, p = 0.23; and Chao1 index: t26 = 1.23, p = 0.23, Fig. 4).
No significant differences were found in community composition, i.e. in beta diversity, between control and manipulator pigs (p = 0.26; PERMANOVA). The visualization of sample similarity with Principal Coordinates Analysis (PCoA; Bray–Curtis dissimilarity) further supported this result (Fig. 5). The control pigs show more variation along the second principal coordinate axis, indicating a more variable gut microbiota.
Sample similarity based on total community composition. Principal coordinates analysis (PCoA) with Bray–Curtis dissimilarity illustrates sample similarity based on amplicon sequence variant (ASV)-level taxonomic profiles (n = 30, PERMANOVA p = 0.26). The lines highlight matched control-manipulator pairs.
Amplicon sequence variant (ASV) was used to distinguish between sequences and to associate sequences with microbes. Of the 20 most important ASVs (detailed list in Supplementary Table S2) with respect to the differences between manipulator and control pigs, most were associated with the families Lachnospiraceae and Ruminococcaceae among the manipulator pigs and with the families Bifidobacteriaceae, Christensenellaceae, and Lachnospiraceae among the control pigs.
Discussion
In this study, pigs at the age of 45 days with manipulative behaviour expressed a higher total abundance of faecal Lactobacillaceae species but a numerically lower alpha diversity. In a study by Rabhi and colleagues13, 13- to 17-week-old pigs of the non-biter/non-bitten negative control group harboured a significantly higher abundance of Lactobacillus than control pigs, whereas Verbeek and colleagues14 did not observe any difference in Lactobacillus between different tail biting phenotypes in their study with pigs aged over nine weeks. As the abundance of Lactobacillaceae tends to build in pigs through late pregnancy towards weaning42, disturbance during this development phase may result in pathology and disease in later life42,43. Shaping of the gastrointestinal microbiome occurs early in life18,20,44,45 and is mainly affected by environmental exposures and diet46,47.
Microorganisms belonging to the families Lachnospiraceae and Ruminococcaceae dominated numerically the faecal microbiota of manipulator pigs. Ruminococcaceae increase in pigs after weaning48 and are linked to plant carbohydrate degradation49. The faecal microbiota of control pigs was characterized by a higher abundance of Bifidobacteriaceae, Christensenellaceae, and Lachnospiraceae. These families have been associated with the preweaning period18,48,49. Bifidobacteria are associated with health benefits50, and they are commonly used as probiotics51. Christensenellaceae have recently moved into the focus of research, as Christensenella minutia has been suggested as a potential probiotic for human health and obesity52. In pigs, Christensenellaceae are linked to fermentation of sugars49.
A narrow range of Lactobacillaceae species has been documented in commercial pigs53, whereas wild boars showed higher alpha diversity than commercial pigs54. This tends to hold true in general; wild Kenyan warthogs scored higher in microbial richness than warthogs kept in captivity (zoo) and higher in alpha diversity than commercial pigs55. This reflects a similar phenomenon in other wild animal species compared with domesticated or semi-domesticated animals54,56,57, indicating higher microbial diversity in non-domesticated animals. In our study, the alpha diversity indices were numerically higher in control pigs, but the difference was not significant. However, our results suggest that a larger sample group could resolve this question. Especially in the female group, for which a subgroup analysis was done (results not presented, p = 0.12), the difference in alpha diversity could potentially become significant by increasing the number of samples.
Sex of the pig proved to be an important factor regarding Lactobacillaceae. A significant interaction with behaviour type was seen for L. amylovorus as well as a tendency for L. reuteri. Sex-related differences regarding tail biting have been shown previously; barrows seem to have a higher frequency of damaged tails than females, but it has not yet been clearly shown that one sex is more prone to perform tail biting8. The influence of sex on microbial composition has been overlooked in previous years, although the topic is gaining in awareness58. Interestingly, Verbeek and others14 found a significant main effect of sex in faecal short-chain fatty acid compositions, which is most probably due to an underlying difference in gastrointestinal microbiota. The effect of sex on microbial diversity was not the focus of this study, but the reasons for differences in faecal Lactobacillaceae composition between females and barrows warrant further research.
A species-specific microbiota composition contributes to animal health and development, with acquisition of a specific selection of microbes resulting in the highest benefit for the host59. Gut microbiota is known to regulate host gene expression60, and therefore, an undisturbed diverse microbiota contributes to richness of metabolites in the host providing several health benefits61. The gut microbiota functions as an endocrine organ62, and it plays a crucial role in the synthesis and regulation of neurotransmitters, e.g. serotonin63. As the evolving microbiota may show low abundance and observed diversity of Lactobacillaceae, a reduced or delayed development of the gut microbiota may be linked to psychological stress64,65,66, potentially causing pig-directed manipulative behaviour. Also, exposure to social stressors has been shown to reduce alpha diversity7, while the gut microbiota can influence the host’s susceptibility to stress67. Tail biting and manipulative behaviour are both suggested to be caused by stress8 and further to create stress in pigs receiving these behaviours3,68. This study, however, does not allow for conclusions on causal relationships or their directions.
Mild manipulation, i.e. no visible harm to tails, in young pigs can turn into damaging tail biting in older pigs10,69. For example, sickness has been shown to lead to manipulative behaviour in pigs69. Nordgreen and others2 have suggested a link between compromised health and damaging behaviour, which may be mediated through the microbiota-gut-brain axis. A high abundance of Lactobacillaceae does not necessarily equal better health, but high diversity and richness have been shown to be associated with better health23 and are suggested to offer the host a more flexible response to environmental changes70. Le Chatelier and colleagues71 reported a low number of gut microbial genes (i.e. a low gene count) to be associated with an unhealthy state, as characterized by higher adiposity, insulin resistance, dyslipidaemia, and low-grade inflammation. Also, Dao and colleagues72 and Cotillard and colleagues73 have confirmed the positive correlations between a high basal gene count, healthier metabolic status, and better outcomes after dietary restriction, supporting the importance of the evaluation of gene richness.
Regarding the limitations of this study, the small sample size remains the biggest limitation. Even though a large number of individually ear-tagged pigs were included to begin with, only 15 manipulators and 15 controls could be reasonably matched. We recognize, that by including as few as 30 pigs into the study, the sample size is quite small yet comparable to similar studies13,14. Further, we compared only two groups in this study. It was not possible for us to form a separate victim-group due to manipulator pigs in many cases being also victims. Recognizing control pigs was also difficult. The behaviour of manipulator and control pigs may not have been sufficiently different from each other to see significant differences in the microbiota. On the other hand, trends observed in this setting could point towards even greater microbial differences during an actual tail-biting episode, which would make it possible to identify more extreme behavioural phenotypes. Secondly, we included only one farm in the study to focus on microbial differences in individuals. This farm was chosen based on frequent tail biting outbreaks in the past. Despite of the history of the farm, no severe lesions were seen on the pigs and mainly ear manipulation was observed and analysed. Thirdly, we acknowledge, that choosing manipulators and controls from the same pens increased the risk of microbes transferring between pen mates since the manipulator-control pairs ate the same feed, were kept under the same conditions, and most of the pairs were within transmission distance. Both Rabhi and colleagues13 and Verbeek and colleagues14 chose biters and controls from different pens. By choosing both manipulator and control primarily form the same pen, our study setup enabled separating the effect of pen and phenotype, which differs from previous studies. Finally, the laboratory methods used may have been a further limiting factor since many of the Lactobacillaceae species could not be identified, instead being included in the total Lactobacillaceae count. The confidence of identification of microbes relies on alignment to a limited set of sequences from culture collections74. Additionally, amplification of the V3-V4 variable region of the 16S rRNA gene does not provide adequate taxonomic resolution to reliably identify bacteria at species or strain level75. Despite of these limiting factors, we were able to pinpoint some significant differences.
In conclusion, these results indicate an association between the faecal microbiota and pig-directed manipulative behaviour while not allowing for conclusions on causality. The difference in microbial composition shows promising trends, warranting further studies. These results suggest that low observed diversity in Lactobacillaceae and the development of manipulative behaviour in pigs may be linked. The link between total faecal microbiota, especially beneficial microbes, and their metabolism, and the development of manipulative behaviour may be better disclosed with a larger sample size and sampling during an ongoing tail biting episode, allowing for the identification of more extreme behavioural phenotypes.
Methods
Ethical statement
The institutional review board of the Regional State Administrative Agency for Southern Finland approved the experiment and the experimental protocol (ESAVI/16950/2018). All experiments were performed in accordance with relevant guidelines and regulations. Reporting in the manuscript follows the ARRIVE guidelines recommendations.
Animals and housing
The study was performed on a commercial farm in South-West Finland in November 2019. The farm was chosen due to a history of frequent tail biting outbreaks. Study pigs were chosen from all piglets (Norwegian Landrace x Norwegian Yorkshire) born on the farm within seven days. Piglets (n = 478) were individually ear-tagged at birth and were visually assigned to three size categories: small, medium, or large. Animal caretakers castrated all male piglets surgically before the age of seven days and relieved their castration pain with intramuscularly injected meloxicam prior to and one day after the castration. No tail docking or teeth clipping was done.
Piglets were housed with their dams in farrowing pens with farrowing crates until weaning at the mean age of 25 (range 23–27) days. Thereafter, piglets were moved to the weaning facilities, with each pen holding 15–22 piglets. A mean of 21 study pigs (range 1–22) were housed in one weaning pen together with non-study pigs in two climate-pens with total space of 10.6 m2. Pens had an elevatable roof over the lying area. Approximately 22% of the pen area was a partly slatted concrete floor. The pigs were provided with one iron chain per pen. Caretakers distributed sawdust, straw, or peat weekly. At least small amounts of these materials were always visible in the pen. Two pens shared a five-m-long feeding trough, and the pigs were liquid fed according to Finnish standards. Pigs had ad libitum access to water from three nipples/pen next to the feeding trough.
Faecal sampling
Faecal samples were collected from study pigs one day before the start of video recording. One researcher held the pigs while another collected a faecal rectal sample with a clean disposable glove and placed it into a faecal sampling tube. Tubes were stored in cooling boxes, moved to – 20 °C within an hour, and transported to – 80 °C at the end of the collection day.
Video recording and behavioural analysis
At the start of the observation period, the mean age of the pigs was 45 (range 43–47) days. Six cameras (IP Camera, IPC-HFW1230S-0280B-BLACK, Zhenjiang Dahua Vision Technology Co., Ltd., Hangzhou, P.R. China) were fixed on the ceiling so that one camera filmed two pens. Recordings were managed from a connected computer with surveillance software (Blue Iris Security, Video Management Software, https://blueirissoftware.com/). Pigs were spray-marked on the back for individual recognition in pens/on video recordings. Markings were reinforced every other day at noon.
The pens were video-recorded for eight consecutive days. Footage from days one, three, and seven was selected for analysis because pigs were marked on these days and to achieve as long a time span as possible. The times of high behavioural activity around feeding were selected for observations. On each of the three days, late afternoon feeding (at approximately 5:30 p.m.) and evening feeding (at approximately 10 p.m.) were observed. These feedings were after the end of the working day of the caretakers, ensuring that the pigs were not disturbed or startled. Observations started one minute before the pigs received feed and lasted for eleven minutes. This adds up to 66 min of observation for each pig. One person analysed all the chosen recordings with a one-zero protocol. After observing the behaviour of each pig for one minute at a time (within the timeframe of eleven minutes/feeding), it was recorded whether the pig performed or received any of the behaviours listed in the ethogram presented in Table 1.
Selection of manipulator and control pigs
The ear-tagged pigs were divided into two rooms within the weaning facilities. A total of 210 pigs were selected from one room for video recording. The selected room contained twelve pens, of which ten pens, containing more than ten study pigs, were chosen. Pigs with one or more records of performing manipulative behaviour on at least two out of the three observation days were defined as manipulators (n = 20). Similarly, pigs with no records of performed or received manipulation were defined as control pigs (n = 21). These pigs were matched to 15 manipulator-control pig pairs as shown in Table 2. Pairs were standardized as far as possible based on sex, birth week size, and pen. At least two of these attributes had to match for the pairs to be included in the study. In the final dataset, we had 17 male and 13 female pigs. Out of these 30 pigs, two were categorized as small, 23 as medium, and five as large at birth.
Lactobacillaceae analysis
Determination of faecal Lactobacillaceae was done as previously described by König and colleagues26. Briefly, faecal samples were thawed on ice and a tenfold dilution series was plated on blood liver (BL) agar. All non-identical colonies were picked from the plate, moved into Gifu Anaerobic Medium Broth, and incubated. Under microscope, non-motile rod-shaped bacteria were streaked on BL agar plates, and their DNA was isolated. Different Lactobacillaceae were identified with a colony PCR, and Lactobacillaceae species were identified with a 16S PCR. The PCR product was purified, the DNA measured and sequenced, and Lactobacillaceae species were identified with the National Center for Biotechnology Information’s (NCBI) Basic Local Alignment Search Tool (BLAST) database with 96% accuracy.
16S rRNA amplicon sequencing
Faecal samples were weighed (0.100–0.125 g) into a two ml screw cap tube and microbiota analysis was performed with InviMag® Stool DNA Kit (Invitek Molecular GmbH, Berlin, Germany) according to the manufacturer’s instructions. A FastPrep®-24 Sample Preparation System (M.P. Biomedicals, Irvine, California, USA), a Heraeus Pico 17 Centrifuge (Thermo Fisher Scientific Ltd., Osterode am Harz, Germany), and a KingFisher™ Purification System Type 700 (Thermo Fisher Scientific Ltd., Vantaa, Finland) were used for the extraction. DNA concentrations of eluates were measured with a NanoDrop 2000 UV–Vis Spectrophotometer (Thermo Fisher Scientific Ltd., Wilmington, Delaware, USA).
DNA concentration was measured using a Qubit® 2.0 Fluorometer (Life Technology, Carlsbad, California, USA) to normalize concentrations to run the libraries. DNA libraries were performed with amplification of the V3-V4 variable region of the 16S rRNA gene with the primers selected from Klindworth and colleagues76, as described previously77. A multiplexing step was conducted with the NextEra XT Index Kit (FC-131-2001) (Illumina, San Diego, California, USA), and DNA quality of the library PCR product was measured with a Bioanalyzer DNA 1000 chip (Agilent Technologies, Santa Clara, California, USA) to verify the size (expected size on a Bioanalyzer trace is ~ 550 bp). The libraries were sequenced using a 2 × 300 bp paired-end run on MiSeq Illumina platform according to the manufacturer’s instructions.
The 16S rRNA amplicon sequences were pre-processed with the DADA278 algorithm (R package dada279), by customizing an established workflow80. The sequences were truncated based on manual investigation of the quality plots (forward 260; reverse 210) and trimmed from the left (base 19). Reads with more than two expected errors were discarded. Otherwise, default settings were used. Chimeric sequences were removed. The ASVs were inferred from the processed sequences. The taxonomic assignment of the ASVs was done against the Silva database (version 138.1)81 downloaded at the URL https://benjjneb.github.io/dada2/training.html (accessed 22 Feb 2022). The abundance tables, taxonomic mappings, and phenotype data were assembled into the TreeSummarizedExperiment data container82.
Data handling and statistical analysis
Lactobacillaceae
For statistical analysis, only clearly identified Lactobacillaceae were considered. Multiple possible species were regarded as unidentified Lactobacillaceae sp. Statistical analyses were done in SPSS (IBM Corp. released 2020. IBM SPSS Statistics for Windows, version 27.0. Armonk, New York, USA). All variables were checked for normal distribution, and only the total number of Lactobacillaceae was found to be normally distributed. A paired samples t-test was performed to check for differences in number of identified Lactobacillaceae species between manipulator and control pigs. To compare the amount of Lactobacillaceae between manipulator and control pigs, linear mixed models (LMM) were conducted. This was, however, possible only for L. amylovorus, L. reuteri, and total Lactobacillaceae since the other identified Lactobacillaceae species were too scarcely represented in the samples. For the LMM, both L. amylovorus and L. reuteri were log10-transformed to achieve close to normal distribution. Initially, all three models included piglet birth size and pen as random factors and sex and status (manipulator or control) as fixed factors. The interaction sex * status was tested for all models but was significant and improved model quality based on AIC value only in models for L. amylovorus and L. reuteri. Model residuals were checked for normal distribution. Differences between manipulator and control pigs in the amount of Lactobacillaceae species present in samples were checked with Mann–Whitney U-test.
Microbial diversity
To assess possible differences in manipulator and control pig microbial diversity the following analyses were performed. Taxonomic profiling data were analysed with the multiassay framework83 using the R package vegan84. A total read count of 0.95 million reads was retrieved for the 30 samples, with an average of 31,819 reads per sample (range 14 917–97 967 reads). These were assigned into 11,056 ASVs (141 singletons) from 36 phyla, 70 classes, 119 orders, 164 families, 351 genera, and 99 species. Alpha diversity was estimated using the Shannon and Chao1 indices. Two-way analysis of variance (ANOVA) models were constructed to compare the status groups and sex, with the main effects of status and sex together with their interaction term. Community composition (beta diversity) was visualized with Principal Coordinates Analysis (PCoA) based on Bray–Curtis dissimilarities. Permutational analysis of variance (PERMANOVA) was used to compare the status groups regarding beta diversity (R function vegan::adonis).
Data availability
The 16S rRNA sequences for this study have been deposited with links to BioProject accession number PRJNA937729 in the NCBI BioProject database: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA937729. The datasets generated during and/or analysed during the current study are available in the Github repository Manipulative_behaviour: https://github.com/emiliakonig/Manipulative_behaviour.
References
Schrøder-Petersen, D. L. & Simonsen, H. B. Tail biting in pigs. Vet. 162, 196–210. https://doi.org/10.1053/tvjl.2001.0605 (2001).
Nordgreen, J. et al. A proposed role for pro-inflammatory cytokines in damaging behaviour in pigs. Front. Vet. Sci. 7, 646. https://doi.org/10.3389/fvets.2020.00646 (2020).
Beattie, V. E. et al. Factors identifying pigs predisposed to tail biting. Anim. Sci. 80, 307–312. https://doi.org/10.1079/ASC40040307 (2005).
Brunberg, E., Wallenbeck, A. & Keeling, L. J. Tail biting in fattening pigs: Associations between frequency of tail biting and other abnormal behaviours. Appl. Anim. Behav. Sci. 133, 18–25. https://doi.org/10.1016/j.applanim.2011.04.019 (2011).
Taylor, N. R., Main, D. C. J., Mendl, M. & Edwards, S. A. Tail-biting: A new perspective. Vet. J. 186, 137–147. https://doi.org/10.1016/j.tvjl.2009.08.028 (2010).
Valros, A. Chapter 5—Tail biting. In Advances in Pig Welfare (ed. Spinka, M.) 137–166 (Woodhead Publishing, 2018).
Kobek-Kjeldager, C., Schönherz, A. A., Canibe, N. & Pedersen, L. J. Diet and microbiota-gut-brain axis in relation to tail biting in pigs: A review. Appl. Anim. Behav. Sci. 246, 105514. https://doi.org/10.1016/j.applanim.2021.105514 (2022).
Edwards, S. & Valros, A. Understanding and preventing tail biting in pigs. In Understanding the Behaviour and Improving the Welfare of Pigs (ed. Edwards, S.) 1–38 (Burleigh Dodds Science Publishing, 2021).
Valros, A., Sali, V., Hälli, O., Saari, S. & Heinonen, M. Does weight matter? Exploring links between birth weight, growth and pig-directed manipulative behaviour in growing-finishing pigs. Appl. Anim. Behav. Sci. 245, 105506. https://doi.org/10.1016/j.applanim.2021.105506 (2021).
Schrøder-Petersen, D. L., Simonsen, H. B. & Lawson, L. G. Tail-in-mouth behaviour among weaner pigs in relation to age, gender and group composition regarding gender. Acta Agric. Scand. A 53, 29–34. https://doi.org/10.1080/09064700310002017 (2003).
van der Eijk, J. A. J. et al. Differences in gut microbiota composition of laying hen lines divergently selected for feather pecking. Poult. Sci. 98, 7009–7021. https://doi.org/10.3382/ps/pez336 (2019).
Brunberg, E. I. et al. Omnivores going astray: A review and new synthesis of abnormal behaviour in pigs and laying hens. Front. Vet. Sci. 3, 57. https://doi.org/10.3389/fvets.2016.00057 (2016).
Rabhi, N. et al. Association between tail-biting and intestinal microbiota composition in pigs. Front. Vet. Sci. 7, 563762. https://doi.org/10.3389/fvets.2020.563762 (2020).
Verbeek, E., Keeling, L., Landberg, R., Lindberg, J. E. & Dicksved, J. The gut microbiota and microbial metabolites are associated with tail biting in pigs. Sci. Rep. 11, 20547. https://doi.org/10.1038/s41598-021-99741-8 (2021).
Qin, Y. & Wade, P. A. Crosstalk between the microbiome and epigenome: Messages from bugs. J. Biochem. 163, 105–112. https://doi.org/10.1093/jb/mvx080 (2018).
Milani, C. et al. The first microbial colonizers of the human gut: Composition, activities and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 81, e00036-e117. https://doi.org/10.1128/MMBR.00036-17 (2017).
Gensollen, T., Iyer, S. S., Kasper, D. L. & Blumberg, R. S. How colonization by microbiota in early life shapes the immune system. Science 352, 539–544. https://doi.org/10.1126/science.aad9378 (2016).
Alipour, M. J. et al. The composition of the perinatal intestinal microbiota in cattle. Sci. Rep. 8, 10437. https://doi.org/10.1038/s41598-018-28733-y (2018).
Funkhouser, L. J. & Bordenstein, S. R. Mom knows best: The universality of maternal microbial transmission. PLoS Biol. 11, e1001631. https://doi.org/10.1371/journal.pbio.1001631 (2013).
Husso, A. et al. The composition of the perinatal intestinal microbiota in horse. Sci. Rep. 10, 441. https://doi.org/10.1038/s41598-019-57003-8 (2020).
Rautava, S., Luoto, R., Salminen, S. & Isolauri, E. Microbial contact during pregnancy, intestinal colonization and human disease. Nat. Rev. Gastroenterol. Hepatol. 9, 656–576. https://doi.org/10.1038/nrgastro.2012.144 (2012).
Liu, H. et al. Microbial and metabolic alterations in gut microbiota of sows during pregnancy and lactation. FASEB J. 33, 4490–4501. https://doi.org/10.1096/fj.201801221RR (2019).
van den Elsen, L. W. J., Garssen, J., Burcelin, R. & Verhasselt, V. Shaping the gut microbiota by breastfeeding: The gateway to allergy prevention?. Front. Pediatr. 7, 47. https://doi.org/10.3389/fped.2019.00047 (2019).
Vilson, Å. et al. Disentangling factors that shape the gut microbiota in German shepherd dogs. PLoS ONE 13, e0193507. https://doi.org/10.1371/journal.pone.0193507 (2018).
Alonge, S., Aiudi, G. G., Lacalandra, G. M., Leoci, R. & Melandri, M. Pre- and probiotics to increase the immune power of colostrum in dogs. Front. Vet. Sci. 7, 570414. https://doi.org/10.3389/fvets.2020.570414 (2020).
König, E. et al. Herd-level and individual differences in fecal lactobacilli dynamics of growing pigs. Animals 11, 113. https://doi.org/10.3390/ani11010113 (2021).
Chaaban, H. et al. Early antibiotic exposure alters intestinal development and increases susceptibility to necrotizing enterocolitis: A mechanistic study. Microorganisms 10, 519. https://doi.org/10.3390/microorganisms10030519 (2022).
Francino, M. P. Antibiotics and the human gut microbiome: Dysbioses and accumulation of resistances. Front. Microbiol. 6, 1543. https://doi.org/10.3389/fmicb.2015.01543 (2016).
van de Wouw, M. et al. Short-chain fatty acids: Microbial metabolites that alleviate stress-induced brain-gut axis alterations. J. Physiol. 596, 4923–4944. https://doi.org/10.1113/JP276431 (2018).
Cryan, J. F. et al. The microbiota-gut-brain axis. Physiol. Rev. 99, 1877–2013. https://doi.org/10.1152/physrev.00018.2018 (2019).
Sampson, T. R. & Mazmanian, Z. K. Control of brain development, function, and behaviour by the microbiome. Cell Host Microbe 17, 565–576. https://doi.org/10.1016/j.chom.2015.04.011 (2015).
Averina, O. V. et al. Bacterial metabolites of human gut microbiota correlating with depression. Int. J. Mol. Sci. 21, 9234. https://doi.org/10.3390/ijms21239234 (2020).
Sylvia, K. E. & Demas, G. E. A gut feeling: Microbiome-brain-immune interactions modulate social and affective behaviours. Horm. Behav. 99, 41–49. https://doi.org/10.1016/j.yhbeh.2018.02.001 (2018).
Frankiensztajn, L. M., Elliot, E. & Koren, O. The microbiota and the hypothalamus-pituitary-adrenocortical (HPA) axis, implications for anxiety and stress disorders. Curr. Opin. Nerobiol. 62, 76–82. https://doi.org/10.1016/j.conb.2019.12.003 (2020).
Yano, J. M. et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276. https://doi.org/10.1016/j.cell.2015.02.047 (2015).
Bacqué-Cazenave, J. et al. Serotonin in animal cognition and behaviour. Int. J. Mol. Sci. 21, 1649. https://doi.org/10.3390/ijms21051649 (2020).
Rosado, B. et al. Blood concentrations of serotonin, cortisol and dehydroepiandrosterone in aggressive dogs. Appl. Anim. Behav. Sci. 123, 124–130. https://doi.org/10.1016/j.applanim.2010.01.009 (2010).
Stracke, J., Otten, W., Tuchscherer, A., Puppe, B. & Düpjan, S. Serotonin depletion induces pessimistic-like behaviour in a cognitive bias paradigm in pigs. Physiol. Behav. 174, 18–26. https://doi.org/10.1016/j.physbeh.2017.02.036 (2017).
Ursinus, W. W., Van Reenen, C. G., Reimert, I. & Bolhuis, E. Tail biting in pigs: Blood serotonin and fearfulness as pieces of the puzzle?. PLoS ONE 9, e107040. https://doi.org/10.1371/journal.pone.0107040 (2014).
Valros, A. et al. Evidence for a link between tail biting and central monoamine metabolism in pigs (Sus scorfa domestica). Physiol. Behav. 143, 151–157. https://doi.org/10.1016/j.physbeh.2015.02.049 (2015).
Czycholl, I. et al. Are biters sick? Health status of tail biters in comparison to control pigs. Porc. Health Manag. 9, 19. https://doi.org/10.1186/s40813-023-00314-0 (2019).
Ji, Y. J. et al. Stages of pregnancy and weaning influence the gut micdrobiota diversity and function of sows. J. Appl. Microbiol. 127, 867–879. https://doi.org/10.1111/jam.14344 (2019).
Hashemi, A., Villa, C. R. & Comelli, E. M. Probiotics in early life: A preventative and treatment approach. Food Funct. 7, 1752–1768. https://doi.org/10.1039/C5FO01148E (2016).
Guard, B. C., Mila, H., Mariani, C., Suchodolski, J. S. & Chastant-Maillard, S. Characterization of the fecal microbiome during neonatal and early pediatric development in puppies. PLoS ONE 12, e01775718. https://doi.org/10.1371/journal.pone.0175718 (2017).
Nuriel-Ohayon, M., Neuman, H. & Koren, O. Microbial changes during pregnancy, birth and infancy. Front. Microbiol. 7, 1031. https://doi.org/10.3389/fmicb.2016.01031 (2016).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. https://doi.org/10.1038/nature12820 (2014).
Gschwendtner, S. et al. Early life determinants induce sustainable changes in the gut microbiome of six-year-old children. Sci. Rep. 9, 12675. https://doi.org/10.1038/s41598-019-49160-7 (2019).
Karasova, D. et al. Development of piglet gut microbiota at the time of weaning influences development of postweaning diarrhea—A field study. Res. Vet. Sci. 135, 59–65. https://doi.org/10.1016/j.rvsc.2020.12.022 (2021).
Ruczizka, I. et al. Early parental administration of ceftiofur has gender-specific short- and log-term effects on the faecal microbiota and growth in pigs from the suckling to growing phase. Animals 10, 17. https://doi.org/10.3390/ani10010017 (2019).
Vlasova, A. N., Kandasamy, S., Chattha, K. S., Rajashekara, G. & Saif, L. J. Comparison of probiotic lactobacilli and bifidobacteria effects, immune responses and rotavirus vaccines and infection in different host species. Vet. Immunol. Immunopathol. 172, 72–84. https://doi.org/10.1016/j.vetimm.2016.01.003 (2016).
Sharma, M., Wasan, A. & Sharma, R. K. Recent developments in probiotics: An emphasis on Bifidobacterium. Food Biosci. 41, 100993. https://doi.org/10.1016/j.fbio.2021.100993 (2021).
Chang, C.-J. et al. Next generation probiotics in disease amelioration. J. Food. Drug. Anal. 27, 615–622. https://doi.org/10.1016/j.jfda.2018.12.011 (2019).
Korhonen, J. M., Sclivagnotis, Y. & von Wright, A. Characterization of dominant cultivable lactobacilli ad their antibiotic resistance profiles from faecal samples of weaning piglets. J. Appl. Microbiol. 103, 2496–2503. https://doi.org/10.1111/j.1365-2672.2007.03483.x (2007).
Huang, J. et al. Isolation, characterization and selection of potential probiotic lactic acid bacteria from feces of wild boar, native pig and commercial pig. Livest. Sci. 237, 104036. https://doi.org/10.1016/j.livsci.2020.104036 (2020).
Correa-Fiz, F. et al. Comparative analysis of the fecal microbiota from different species of domesticated and wild suids. Sci. Rep. 9, 13616. https://doi.org/10.1038/s41598-019-49897-1 (2019).
Ang, L. et al. Gut microbiome characteristics in feral and domesticated horses from different geographic locations. Commun. Biol. 5, 172. https://doi.org/10.1038/s42003-022-03116-2 (2022).
Gao, H. et al. Comparison of the gut microbiota composition between the wild and captive Tibetan wild ass (Equus kiang). J. Appl. Microbiol. 126, 1869–1878. https://doi.org/10.1111/jam.14240 (2019).
Kim, Y. S., Unno, T., Kim, B.-Y. & Park, M.-S. Sex differences in gut microbiota. World J. Men’s Health 38, 48–60. https://doi.org/10.5534/wjmh.190009 (2020).
Frese, S. A. et al. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. 7, e1001314. https://doi.org/10.1371/journal.pgen.1001314 (2011).
Richards, A. L. et al. Gut microbiota has a widespread and modifiable effect on host gene regulation. mSystems 4, e00323-18. https://doi.org/10.1128/mSystems.00323-18 (2019).
Nichols, R. G. & Davenport, E. R. The relationship between the gut microbiome and host gene expression: A review. Hum. Genet. 140, 747–760. https://doi.org/10.1007/s00439-020-02237-0 (2021).
Rastelli, M., Cani, P. D. & Knauf, C. The gut microbiome influences host endocrine functions. Endocr. Rev. 40, 1271–1284. https://doi.org/10.1210/er.2018-00280 (2019).
Hata, T. et al. Regulation of gut luminal serotonin by commensal microbiota in mice. PLoS ONE 12, e0180745. https://doi.org/10.1371/journal.pone.0180745 (2017).
Bailey, M. T. et al. Exposure to a social stressor alters the structure of the intestinal microbiota: Implications for stressor-induced immunomodulation. Brain Behav. Immun. 25, 397–407. https://doi.org/10.1016/j.bbi.2010.10.023 (2011).
Marin, I. A. et al. Microbiota alteration is associated with the development of stress-induced despair behavior. Sci. Rep. 7, 43859. https://doi.org/10.1038/srep43859 (2017).
Yu, M. et al. Variations in gut microbiota and fecal metabolic phenotype associated with depression by 16S rRNA gene sequencing and LC/MS-based metabolomics. J. Pharm. Biomed. Anal. 138, 231–239. https://doi.org/10.1016/j.jpba.2017.02.008 (2017).
Nowland, T. L., Plush, K. J., Barton, M. & Kirkowood, R. N. Development and function of the intestinal microbiome and potential implications for pig production. Animals 9, 76. https://doi.org/10.3390/ani9030076 (2019).
Valros, A. et al. Physiological indicators of stress and meat and carcass characteristics in tail bitten slaughter pigs. Acta Vet. Scand. 55, 75. https://doi.org/10.1186/1751-0147-55-75 (2013).
Munsterhjelm, C. et al. Sick and grumpy: Changes in social behaviour after a controlled immune stimulation in group-housed gilts. Physiol. Behav. 198, 76–83. https://doi.org/10.1016/j.physbeh.2018.09.018 (2019).
Voolstra, C. R. & Ziegler, M. Adaptationg with microbial help: Microbiome flexibility facilitates raid responses to environmental change. BioEssays 42, 2000004. https://doi.org/10.1002/bies.202000004 (2020).
Le Chatelier, E. et al. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. https://doi.org/10.1038/nature12506 (2013).
Dao, M. C. et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: Relationship with gut microbiome richness and ecology. Gut 65, 426–436. https://doi.org/10.1136/gutjnl-2014-308778 (2016).
Cotillard, A. et al. Dietary intervention impact on gut microbial gene richness. Nature 500, 585–588. https://doi.org/10.1038/nature12480 (2013).
Srinivasan, R. et al. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS ONE 10, e0117617. https://doi.org/10.1371/journal.pone.0117617 (2015).
Johnson, J. S. et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 10, 5029. https://doi.org/10.1038/s41467-019-13036-1 (2019).
Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. Spec. Publ. 41, e1. https://doi.org/10.1093/nar/gks808 (2013).
Gómez-Gallego, C. et al. Glyphosate-based herbicide affects the composition of microbes associated with Colorado potato beetle (Leptinotarsa decemlineata). FEMS Microbiol. Lett. 367, fnaa050. https://doi.org/10.1093/femsle/fnaa050 (2020).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. https://doi.org/10.1038/nmeth.3869 (2016).
Callahan, B., McMurdie, P. & Holmes, S. Dada2: Accurate, high-resolution sample inference from amplicon sequencing data. http://benjjneb.github.io/dada2/ (2020).
Callahan, B. J., Sankaran, K., Fukuyama, J. A., McMurdie, P. J. & Holmes, S. P. Bioconductor workflow for microbiome data analysis: From raw reads to community analyses [Version 2; Peer Review: 3 Approved]. F1000Research 5, 1492. https://doi.org/10.12688/f1000research.8986.2 (2016).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 41, D590–D596. https://doi.org/10.1093/nar/gks1219 (2013).
Huang, R. et al. TreeSummarizedExperiment: A S4 Class for data with hierarchical structure [version 2; peer review; 3 approved]. F1000Research 9, 1246. https://doi.org/10.12688/f1000research.26669.2 (2021).
Lahti, L. et al. Orchestrating Microbiome Analysis with Bioconductor [Beta Version]. microbiome.github.io/OMA/ (2022).
Oksanen, J.F. et al. Vegan: Community Ecology Package. https://CRAN.R-project.org/package=vegan (2022).
Acknowledgements
Open access funded by Helsinki University Library. This work was supported by the Ministry of Agriculture and Forestry of Finland [grant no. Makera 504/03.01.02/2018], A-Rehu Ltd., Vetcare Ltd., and the Finnish Foundation of Veterinary Research. LL was supported by the Academy of Finland (decision no. 295741). The authors thank the farm for agreeing to the video-recording of their pigs. The authors also thank Sini Marttila for indispensable help on the farm and Jeferson Delgado Florez for valuable reviewing and comments.
Author information
Authors and Affiliations
Contributions
E.K., V.P., S.B., S.S., M.H., and A.V. conceived the experiment. E.K., S.B., V.H., M.N., M.H., and A.V. conducted the experiment. E.K., P.H., S.K., J.R., M.C.C., T.B., L.L., J.J., S.B., S.S., M.H., and A.V. analysed the results. All authors reviewed the manuscript and approved the final version.
Corresponding author
Ethics declarations
Competing interests
At the time this study was conducted, S.B. was an employee of Vetcare Ltd. Other authors do not have any competing interests to declare.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
König, E., Heponiemi, P., Kivinen, S. et al. Fewer culturable Lactobacillaceae species identified in faecal samples of pigs performing manipulative behaviour. Sci Rep 14, 132 (2024). https://doi.org/10.1038/s41598-023-50791-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-50791-0