Abstract
One of the main challenges in coral reef conservation and restoration is the identification of coral populations resilient under global warming. Seascape genomics is a powerful tool to uncover genetic markers potentially involved in heat tolerance among large populations without prior information on phenotypes. Here, we aimed to provide first insights on the role of candidate heat associated loci identified using seascape genomics in driving the phenotypic response of Acropora millepora from New Caledonia to thermal stress. We subjected 7 colonies to a long-term ex-situ heat stress assay (4 °C above the maximum monthly mean) and investigated their physiological response along with their Symbiodiniaceae communities and genotypes. Despite sharing similar thermal histories and associated symbionts, these conspecific individuals differed greatly in their tolerance to heat stress. More importantly, the clustering of individuals based on their genotype at heat-associated loci matched the phenotypic variation in heat tolerance. Colonies that sustained on average lower mortality, higher Symbiodiniaceae/chlorophyll concentrations and photosynthetic efficiency under prolonged heat stress were also the closest based on their genotypes, although the low sample size prevented testing loci predictive accuracy. Together these preliminary results support the relevance of coupling seascape genomics and long-term heat stress experiments in the future, to evaluate the effect size of candidate heat associated loci and pave the way for genomic predictive models of corals heat tolerance.
Similar content being viewed by others
Introduction
Coral reefs are considered one of the most vulnerable ecosystems to global warming1 with around 50% of global cover already lost since 19502,3,4,5,6. The exacerbated decline in global coral cover in the recent decade has been mainly attributed to mass bleaching triggered by abnormally high sea-water temperatures5,6. Coral bleaching occurs when symbiotic microalgae (Symbiodiniaceae) living within the host are expelled or digested leading to nutritional deprivation7,8 which can ultimately lead to coral death9. Increasingly frequent and long marine heatwaves10 are shortening return times between bleaching events11. Under moderate emissions scenarios (RCP 4.5) bleaching events could occur annually in > 75% of global coral reefs before 207012. Therefore, the Intergovernmental Panel on Climate Change predicts that 70–90% of global coral cover will be lost by 2100 under a 1.5 °C warming scenario13. However, these predictions do not generally take into account contrasted heat stress responses among geographical regions14, species15, morphotypes16, thermal history17 and environmental conditions (e.g., hydrodynamic flow18, nutrient enrichment19,20 and light irradiance21). In addition, associated Symbiodiniaceae22,23 and bacterial communities24,25 can affect the thermal and bleaching tolerance of coral colonies.
Heat tolerance has also been shown to vary among conspecific colonies from similar habitats26,27,28,29, suggesting that individual effects can enhance or alter the resilience of populations under warmer conditions. Such differences may be due to acclimatization through phenotypic plasticity (e.g., changes in gene expression30,31 or Symbiodiniaceae communities32,33), transgenerational effects (e.g., epigenetic modifications34 and symbiont transmission35) or standing genetic variation in the host. Corals heat tolerance has been found to be a heritable trait with a polygenic basis28,36,37,38,39,40. Despite evidences for contemporary increases in coral heat tolerance41,42,43, how much and where genetic adaptation can keep pace with global warming is yet to be determined in most reef systems. It depends in particular on several parameters that are unknown in most taxa such as heat tolerance heritability44,45,46, selection differential45, covariance between traits (e.g., growth and symbiont retention)47 and potential tradeoffs48,49. Identifying genetic markers involved in heat tolerance and understanding their distribution through time and space is therefore crucial to predict the future of coral reefs50,51,52,53,54 as well as targeting tolerant populations and individuals for conservation55,56,57,58 and restoration46,59,60.
In the last decade, studies have begun to identify host genomic markers associated with corals heat tolerance. Early studies identified candidate loci using transcriptomic signatures in adults31,61,62 and larvae37, or genomic associations with larvae survival under thermal stress45,46. Other studies identified genomic regions showing signals of population differentiation between distinct thermal environments39,40,63. Molecular pathways associated with candidate genes or genomic regions included immune system response, inflammatory and antioxidant response, calcium homeostasis, apoptotic processes, mitochondrial metabolism and nitrogen metabolism. However, most of these studies identified candidate loci using a limited number of individuals originating from a few reefs and without a measurement of individual-level heat tolerance phenotype. This has hindered their application to the screening of tolerant individuals in natural populations and called for new methods to investigate simultaneously genotypes and phenotypic response over larger spatial scales and genomic coverage. Genome wide association studies (GWAS) have been extensively used in the last decade to predict phenotypic traits of individuals given the information about their genotypes64. The first study using this method in corals identified genomic regions associated with bleaching susceptibility in Acropora millepora from 12 populations of the great barrier reef (GBR)67. Despite no individual loci reaching statistical significance due to limited sample size, a predictive model incorporating symbiont identity, environmental data and polygenic scores was able to predict ~ 62% of the variance in bleaching intensity.
Other population genetic methods such as seascape genomics can be used to identify candidate heat associated loci (HAL) without prior information on phenotypes68. In this approach, HAL are detected through correlation tests between allele frequencies and environmental gradients while accounting for the genetic structure of natural populations69. This method was used in a previous study to reveal candidate heat associated loci in A. millepora using 167 individuals sampled across 20 reefs of New Caledonia (Southwest Pacific)70. A. millepora is a model species for studying thermal tolerance as it is considered highly vulnerable to global warming71, has a wide distribution area within the Indo-Pacific region72 and extensive genetic information is available including a chromosome-level reference genome67,73. In addition, several studies have investigated its response to heat stress using physiological measurements, metabarcoding, metabolomics and transcriptomics74,75,76.
In this study we aimed to conduct a preliminary assessment of the relevance of candidate HAL identified using the New Caledonian seascape genomics results70 to explain differences in thermal tolerance between individuals. We subjected 7 colonies originating from the same environment and harboring similar life history traits to a long-term heat stress experiment. Heat-stress has been shown to trigger various physiological and molecular cascades (e.g., generation of reactive species, nutrient cycling and calcium homeostasis impairment) which can result in different endpoints such as host cell death, symbiont expulsion or symbiont degradation7,8. Therefore we used multiple physiological measurements at the end of the experiment to decipher different types of phenotypic response among colonies. Then we investigated a statistical approach to assess the correlation between genotypes at the candidate HAL and phenotypes in the case of limited sample size.
Material & methods
Sampling site, coral collection and fragmentation
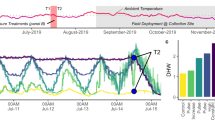
Seven A. millepora colonies were collected at Snark Reef (−22.4388, 166.4292, Supplementary Fig. 1), New Caledonia in February 2021 (permit number: 340–2021/ARR/DDDT). This reef was chosen due to its high bleaching alert frequency since 1981 compared to all New Caledonia reefs (BAF; > 95% percentile; Supplementary Fig. 2), associated with high allelic diversity at HAL70. This was meant to increase the likelihood that genotypes would display different levels of heat tolerance while originating from the same reef. All colonies were collected at a depth ranging between 2 and 3 m, and were located at least 5 m from each other to avoid genotype duplicates. A small fragment (< 1cm3) of each colony was directly stored in 80% ethanol for DNA extraction (Fig. 1). Colonies were maintained in a coral garden in St-Marie Bay (S −22°1717, E 166°2850) for ~ 5 months before performing the experiment. In July 2021, each colony was fragmented to 40 nubbins (except for colonies 13 n = 37, 1 and 3 n = 33) and transferred to experimental tanks at the Aquarium des Lagons Nouméa. Nubbins were randomly distributed throughout four ~ 190 L tanks (2 tanks per treatment, 70 nubbins per tank, 10 nubbins per colony, n = 7 colonies, Supplementary Tab. 1). Of note, colonies 1 and 3 were fragmented earlier in March 2021 and therefore had a longer post-fragmentation acclimation period.
Experimental design and methods to assess the relevance of molecular markers identified through seascape genomics for Acropora millepora heat tolerance. A small sample (< 1cm3) was taken from each colony at the time of collection and stored in ethanol 80% for further ITS2/psbAncr profiling and DART sequencing. Colonies were fragmented in ~ 40 nubbins and randomly split between control and stress treatments (2 tanks/treatment) Visual scores and Fv/Fm were monitored throughout the experiment (2–3 days interval). At the end of the experiment 3 nubbins/colony/treatment were frozen to measure symbiont density and total chlorophyll content.
Experimental design
The experimental design and tank conditions were chosen in line with the general framework suggested by Grottoli et al.77 to increase comparability between studies (Fig. 1). The system consisted of four replicated ~ 190 L tanks supplied with freshly filtered seawater pumped in nearshore waters using 5 µm cartridge filters and 1 L.min-1 flow rate (corresponding to a ~ 7.5 × daily turnover). Temperature control in each tank was achieved using Eliwell EWDR983 controllers connected thermal probes and heating resistors (HT300, Tetra and Pro Temp s300, JBL) in upstream homogenization tanks. Current pumps (WP100, Tetra) maintained a 5–20 cm.s−1 flow rate and 200W LEDs (Maxspect RSX 200) supplied ~ 250 µmol photons m−2.s−1 with a 11:30/13:30 light regime similar to in situ conditions. Temperatures inside the tanks were recorded at 15 min interval with Hobo loggers (Onset, 2 per tank) and pH and salinity were measured daily using a Pro Plus multiparameter meter (YSI). Coral nubbins were fed daily with 5 mL of Artemia larvae cultures (Day 2) per tank (~ 350 larvae/mL) to avoid heterotrophic starvation caused by the water filtration and additional oxygen was provided with air pumps.
Heat stress experiment
Once distributed in the tanks, all nubbins underwent a 2 weeks progressive acclimation to control conditions during which the light intensity was increased from 100 to 250 µmol photons m−2.s−1 and the temperature raised from the lagoon temperature to 27 °C, the Maximum Monthly Mean (MMM) of Snark Reef site (day 0–14). As 1–4 °C above the MMM are expected to elicit a bleaching response within a few weeks77, the water temperature of two tanks was then increased (at a rate of 0.5 °C/day) to 31 °C (MMM + 4) in two steps: from MMM to MMM + 2 (29 °C, day 15–29, and, after 14 days, from MMM + 2 to MMM + 4. MMM + 4 was maintained for 10 days (day 36–46, Fig. 2b).
Coral photosynthetic performances throughout the experiment. (a) Average nubbins maximum photosynthetic efficiency (Fv/Fm) in each treatment: Control (blue) and Stress (red). Error bars represent standard deviation. Asterisks indicate significant differences between treatments (Bonferroni corrections, *p < 0.005, **p < 1e-11, ***p < 1e-22). (b) Temperature profiles of the 4 tanks: Stress (1,4, orange and red), Control (2–3, pale and dark blue). Dashed vertical lines indicate time points where the temperature was increased to MMM +2 and MMM +4.
Visual assessment of coral health
Coral health was monitored throughout the experiment using visual observations. Bleaching scores were assigned using the same criteria as Santoro et al.78: H = healthy, P = paling (< 10% of the nubbin with pigment loss), B = bleached (10% < X < 80%), W = bone white (> 80% of the nubbin white) and D = dead (0% of living tissue). Daily pictures of each nubbin were taken with an underwater camera (Nikon Coolpix W300) to assign bleaching or death dates and coral fragments were observed under a binocular magnifier to assign the correct category when needed. Dead nubbins were removed from the tanks to avoid any pathogen proliferation and contamination of healthy nubbins.
Photosynthetic performance, Symbiodiniaceae density and chlorophyll content
Maximum photosynthetic yield (Fv/Fm) was assessed via pulse-amplitude modulation (PAM) fluorometry at a 2–3 days’ interval during the course of the experiment. This physiological variable is a common proxy of bleaching onset77 and is non-invasive which allows the quantification of holobiont health across time and temperature exposure. Measurements were carried out at least 30 min after artificial sunset to ensure dark-adaptation, using an Aquapen-P AP-110P (photon systems instruments) with a blue measuring light (455 nm) and a saturation pulse intensity of 1500 µmol m−2 s−1.
At the end of the experiment, 6 random nubbins/colony (3 from control and 3 from stress treatments) were frozen for symbiont density and chlorophyll content quantification, which are more accurate but destructive measurements of bleaching intensity, except for colony 10 that did not have any living nubbins left in the stress treatment (36 nubbins in total). Coral tissues were separated from the skeleton using the air pick method79 and Symbiodiniaceae cells released in 0.1 µm filtered seawater through manual homogenization with a potter tissue grinder. Symbiodiniaceae cells were counted using a Malassez cell under a microscope (Leica DM2000, 200 × magnification, 3 replicates per sample, 4 sets of 16 squares per replicate). A 10 mL subsample of the tissue homogenate was centrifuged (6000 g, 10 min) and the supernatant removed. The pellet containing pigments was extracted in 99% acetone (24 h, 4 °C). The total concentration in chlorophyll a (Chla) and chlorophyll c2 (Chlc2) were calculated from absorbance measurements taken with a spectrophotometer (UV-Visible Evolution 201, THERMO, 2014) according to Jeffrey, S. t. and Humphrey, G equations80. Chlorophyll content and Symbiodiniaceae density were normalized to seawater volume and nubbins’ surface area estimated with the paraffine method which is considered to match the accuracy of computer tomography (CT) for the Acropora genus81.
DNA extraction, amplification and sequencing
Bulk DNA was extracted using a chloroform-CTAB protocol82 and sent to diversity arrays technology (Canberra, Australia) for medium density sequencing (1.2 M reads). The DArT -sequencing method (DArT-seq)83 screens thousands of SNP markers across hypomethylated regions of the genome. Library preparation and sequencing were performed using the same protocol and sequencer as described in Selmoni et al. (2021)70.
Additionally, DNA samples were sent for Symbiodiniaceae nuclear intergenic region ITS2 and chloroplastic non coding region psbAncr84 markers amplification (Herculase II Fusion DNA Polymerase Nextera XT Index Kit V2) and Illumina sequencing at Macrogen (Seoul, Korea). ITS2 has a broad phylogenic application range while psbAncr can assess relatedness among close lineages85. The ITS2 hypervariable region was amplified using the forward primer SYM_VAR_5.8S 5’-GAATTGCAGAACTCCGTGAACC and reverse SYM_VAR_REV CGGGTTCWCTTGTYTGACTTCATGC-3′86. The psbAncr from 4 colonies (1,9,11,12) was amplified using the forward 7.4-Forw 5′-GCATGAAAGAAATGCACACAACTTCCC-3′ and reverse 7.8-Rev 5′-GGTTCTCTTATTCCATCAATATCTACTG-3′ primers84.
ITS2 and psbAncr profiling pipeline
Sequencing of the DNA sample from all colonies yielded 160,806–184,696 reads per sample for ITS2 and 109,195–408,758 reads per sample for psbAncr. Adapters were trimmed from ITS2 and psbAncr sequences using Cutadapat v3.187. The Symportal (https://symportal.org) analytical framework was used to analyze Symbiodiniaceae ITS2 sequence data88. ITS2 sequence variants and ITS2 type profiles abundance count tables outputted by the Symportal analysis were used to plot the results. For psbAncr nucleotide sequences, the dada2 ITS pipeline89 was adapted to focus on forward reads only as they had overall higher quality scores and reference sequences are mostly available in the forward direction at this marker. After filtration, dereplication and chimera removal, 784,670 reads were grouped in 50 amplicon sequence variants (ASVs)90 at 100% identity and blasted against NCBI nucleotide database91. Phyloseq package was used for further handling of the data and computation of variants relative abundances92. ASVs that did not match to Symbiodiniaceae reference sequences (19 ASVs, ~ 1.6% of total reads) and low abundant ASVs (4 ASVs, total reads < 10) were discarded. The final set of 27 ASVs was used for phylogenic reconstruction together with Cladocopium forward psbAncr reference sequences from a recent study93. Sequences were aligned by ClustalW and manually edited in MEGA X software94. Jukes-Cantor model with gamma distributed substitution rates (JC + G) was chosen based on aikake information criterion (AIC) using NJ tree reference under automatic model selection in MEGA. A maximum-likelihood (ML) phylogenic tree using this substitution model was reconstructed in MEGA and statistical significance of internal branching assessed using 1000 bootstrap replicates.
Seascape genomics and host genotyping
Candidate markers for heat adaptation were previously identified using a seascape genomics approach based on DArT-seq genotyping of 167 A. millepora colonies from 20 New Caledonian reefs70. This GEAs revealed genomic regions that may be involved in heat stress adaptation by associating SNP frequencies to environmental gradients in mixed models accounting for neutral genetic structure of the population. The DART-seq reads of the new 7 colonies were aligned against the same reference genome (A.millepora v2 chromosome-level assembly67) using DArTSoft14 algorithm to call the same SNPs as in the previous study. The DART-seq pipeline resulted in 55,898 bi-allelic SNPs and the average number of missing genotypes per SNP was 2.3. These SNPs were filtered out to only retain those genotyped for all of the 7 colonies and those that were shared with the 57,374 high quality SNPs obtained in the previous study after filtration of high missing rates (> 50%), rare alleles (minor allele frequency < 5%) and highly frequent genotypes (major genotype frequency > 95%)70. The final genome matrix was composed of 16,504 high quality SNPs. In addition, SNPs had been previously annotated by performing a similarity search of the protein sequence coded by their closest gene against Uniprot/swissprot databases 70.
Statistical analysis
All statistical analysis were conducted using R v4.0.495. The effect of temperature treatment on maximum photosynthetic efficiency throughout the experiment was first investigated using a linear mixed effect model with package lme496. A first model incorporated treatment and time interaction as a fixed effect and random intercepts for each colony, tank and fragmentation date. A second model included the interaction with colony in the fixed effect to investigate potential differences in heat response between colonies over time. Diagnostic plots were inspected to confirm that hypothesis on residuals homoscedasticity and normality were met, and 95% bootstrap confidence intervals were derived for fixed effects. Marginal temperature and colony effects were analyzed at each time point using package emmeans97 and p-values adjusted using Bonferroni correction. Physiological measurements at the end of the MMM + 4 stress period: Chla, Chlc2, symbiont density (n = 3 nubbins/colony/treatment) were compared between colonies and treatments using ANOVA, including fragmentation date as a cofactor. Tukey Post-hoc tests were used to make multiple comparisons between groups as Levene’s test confirmed homogeneity of variance. The accuracy of nubbins’ visual scores was assessed using Principal Component Analysis (PCA) on Chla, Chlc2, symbiont density and Fv/Fm measurements of these 36 nubbins to visualize whether or not bleaching categories correctly reflected their physiological state. A bleaching mortality index was computed to summarize colonies heat tolerance across replicate fragments using a similar approach as Humanes et al.29:
where H,P,B,W,D are the proportion of fragments assigned to each category. Finally a PCA was performed on all phenotypic variables from the 7 colonies in the stress treatment at the end of the experiment (BMI, Fv/Fm, Chlorophyll content and symbiont density) to provide an overview of the host and symbiont heat stress responses from each colony. The first two principal components were used to compute a phenotype dissimilarity matrix that was further tested for correlation with the genotype (see below).
We sought to assess the relevance of molecular markers identified through seascape genomics in explaining bleaching tolerance of these 7 colonies (method summarized in Supplementary Fig. 3). For this purpose, we used Mantel tests to investigate correlations between the phenotype dissimilarity matrix and a dissimilarity matrix based on their genotypes. The previous seascape genomic approach predicts for each SNP an association q-value with two Bleaching Alert Frequency variables computed from satellite data at 1 km and 5 km resolution (BAF, corresponding to the frequency of daily degree heating weeks > 070,98). The genotype dissimilarity matrix was computed using a restricted set of multiple SNPs with a q-value below a specific threshold for at least one BAF variable. The effect of genotype was thus tested as a whole rather than separately for each individual SNP because of the low sample size. Two dissimilarity metrics were computed from the genotypes: allelic dissimilarity (or hamming distance) and euclidean distance based on associated BAF. To compute the second distance, for each SNP, the 167 A. millepora colonies from the previous study were split based on their genotypes (0: homozygote reference, 1: heterozygote, 2:homozygote alternative) and the median BAF of their original reef computed for each of the three groups (Supplementary Fig. 3). The euclidean distance between two individuals was computed based on the BAF associated to each of their respective genotypes. We repeated the computation for 100 genotype dissimilarity matrices, each based on SNPs with an association q-value below thresholds ranging between 0.001 (8 SNPs, strongly associated with BAF) and 0.1 (153 SNPs, some of which may not be associated to heat stress response). For each genotype dissimilarity matrix, Mantel tests were used to assess their correlation with the phenotype dissimilarity matrix. As a control, genotype dissimilarity matrices were also computed from random samples of SNPs across the genome (5–120/16 504 SNPs corresponding to the size of the genotype matrix for q.values of 0.001–0.1, 1000 replications per sampling size). A hierachical clustering of the 7 colonies was performed on the 22 SNPs—for which the Mantel test significance was the strongest—using package pvclust99 (average, allelic dissimilarity, 100 bootstrap replications). Finally, we assessed the predictability of phenotypes through polygenic scores computed from these 22 SNPs as 1) the fraction of heat-adapted genotypes (i.e., variants associated with the highest BAF) and 2) the sum of BAF associated to each genotype (Supplementary Fig. 4). Figures and graphs were made using R package ggplot2100.
Results
Overall heat stress response throughout the experiment
Photosynthetic performances throughout the experiment showed an interaction effect of treatment and time (Fig. 2). Fv/Fm decreased by as much as 22 ± 17% of its initial value in both treatments at the beginning of the acclimation period, potentially as a result of tank transfer, and was restored to 92% of its initial value after 10 days. The effect of temperature on Fv/Fm was negligible during the acclimation period as well as during two weeks at MMM + 2 but became apparent 3 days after reaching MMM + 4, with a gradual 8–42% reduction in the stress treatment throughout days 40–45 (β = −0.03–−0.22, linear mixed effect model, Supplementary Tab. 2). This significant decrease in Fv/Fm was consistent among colonies (p.adj < 0.05, Bonferroni correction, Supplementary Fig. 5, Supplementary Tab. 3) although it was artificially attenuated by the removal of dead nubbins in colonies 10–11 at day 45. Fragmentation date and tank had minor and small effects respectively on Fv/Fm throughout the experiment (explained 0.48 and 7.5% of random variance respectively). Visual observations of coral bleaching and mortality of the nubbins were on par with photosynthetic measurements with no visible differences between controls and treatments during the acclimation period and after two weeks at MMM + 2. Of note, a few nubbins showed sign of tissue loss both in control (colonies 1,3, N = 4) and stress treatment (colonies 11,13 N = 3) and were removed from the tanks. Visual observations of coral bleaching and mortality in the stress treatment mainly occurred during the MMM + 4 window concomitantly with the decline in Fv/Fm.
Visual classification of nubbins health
A PCA was performed on physiological parameters (Fv/Fm, symbiont density, chlorophyll content) from a subset of 36 nubbins at the end of the experiment. Bleaching scores clearly separated along the first component (82.8% explained variance, Supplementary Fig. 6) that defined a size effect. Therefore, the classification of nubbins by their visual scores appears to be a reliable proxy for their physiological state.
Overall heat stress effect at the end of the experiment
Heat stress triggered a strong decline in all physiological measurements measured on 6 colonies (colony 10 did not have enough living nubbins) at the end of the experiment (day 45, ANOVA, Supplementary Tab. 4). In the control treatment, the majority of nubbins remained healthy with symbiont density ranging between 0.5 × 105 and 2.6 × 105 cells.cm-2, Fv/Fm ~ 0.6, chlorophyll a content ranging between 2.5 and 11.0 µg.cm−2 and chlorophyll c2 between 0.23 and 1.54 µg.cm−2 (Fig. 3 and Supplementary Fig. 7). In the stress treatment, nubbins underwent a relative drop of 71 ± 16 in symbiont density (p = 8.4e−7), 76 ± 13% in chlorophyll a (p = 2.6e−6), 52 ± 24% in chlorophyll c2 (p = 9.1e−4) and 42 ± 7% in Fv/Fm (p.adj = 5.2e−23, Bonferroni correction) compared to the control treatment. Fragmentation date did not affect any of the physiological parameters at the end of the experiment (ANOVA, p > 0.05, Supplementary Tab. 4). Overall, the Bleaching Mortality Index was 0.154 in the control treatment and 0.558 in the stress treatment (Fig. 4b), reflecting the global decline in nubbins health under MMM + 4 (e.g., mortality rates of ~ 7% in the control treatment and ~ 27% in the stress treatment).
Symbiodiniaceae physiological measurements at the end of the experiment. (a) Maximum photosynthetic yield (Fv/Fm). Horizontal bars show significant differences between groups with Bonferroni corrections for multiple comparisons and p.values and significance are indicated above. Each point corresponds to measurement of living individual fragments at the end of the experiment. (b) Symbiodiniaceae cells density. Horizontal bars show significant differences between groups, multiple comparisons adjusted with Tukeypost-hoc tests and p.values and significance are indicated above. Each point corresponds to individual fragments average symbiont densities across three replicate aliquots.
Visual health of 7 colonies exposed to a long-term heat stress experiment. (a) Proportion of bleaching categories (visual score) per colony and treatment at the end of the experiment. Nubbins were categorized as healthy, paling (> 10% of the nubbin with pigment loss), bleached, bone white (> 80% of the nubbin white) or dead (0% of living tissue). Dead nubbins were removed from the tanks to avoid pathogen contamination. (b) Individual Bleaching Mortality index per colony and treatment computed from proportion of bleaching categories among fragments of group.
Variation in phenotypic response among colonies
While the control treatment did not show any difference in visual scores (Fig. 4a) or physiological parameters between colonies (ANOVA, p.adj > 0.05, Supplementary Tab. 5), the stress treatment elicited a global decline in nubbins health with substantial variation in colonies response to thermal stress. In the stress treatment, colony 11 showed an earlier decrease (from day 38) in Fv/Fm than other colonies although not significant at 0.05 level (linear mixed effect models, Bonferroni correction, Supplementary Tab. 3) followed by colony 10 (from day 43) and colonies 1,3,9 (day 45). Colony 12 maintained the highest Fv/Fm until the end of the experiment, being 25% higher than colony 10 (p.adj = 0.015) and 11 (p.adj = 9.9e-5) at day 45. Of note statistical power of colony 10 comparisons to other colonies was reduced because most nubbins underwent rapid mortality and were removed from the tanks. Differential response to heat stress held at day 45, with colonies 12,13 showing on average 40% higher chlorophyll a, 43% higher chlorophyll c2 content, 34% higher symbiont cell density than colonies 1,3,9 and respectively 81, 86 and 96% higher levels than colony 11 (although not significant at 0.05 level due to low number of replicates, Supplementary Tab. 5). Bleaching mortality index was also the lowest for colonies 12,13 (~ 0.31), intermediate for colonies1,3,9 (~ 0.47) and the highest for colonies 10,11 (~ 0.94, Fig. 4b). The first component of a PCA (90.6% explained variance, Fig. 5) performed on these 5 phenotype variables (BMI, symbiont density, chlorophyll a and c2 content, Fv/Fm) summarized this size effect, separating the 7 colonies by low (10,11), medium (1,3,9) and high (12,13) thermal tolerance. While all physiological parameters showed consistent decline between clusters, the second component of the PCA (5.5% explained variance) separated colony 13 with greater density of symbionts and colony 12 with higher photosynthetic efficiency which suggests that different physiological mechanisms may be involved in their respective tolerance.
PCA component analysis (PCA) on phenotypic measurements of the 7 colonies at the end of the experiment, after being exposed to MMM +4 °C for 10 days. Component loads projected on the first two components are depicted in arrow. Numbers in parentheses represent the proportion of variance explained by that principal component. The barplot on the right-hand indicates the contribution of each phenotypic measurements to the first principal component.
Endosymbiont communities
All colonies shared ITS2 type profiles representing symbionts from the Cladocopium genus (Fig. 6a). Symbiodiniaceae communities were predominated by variants C50b (~ 53% of total reads) followed by C3 (~ 12%), C3bo (7%), C3k (3%), C3bp (3%) and C50aq (< 2%). At the exception of colony 9, all colonies showed ITS2 type profile dominated by variant C50b (C50b-C3-C3bm-C50f, C50b/C3, C50b/C3k-C3bo-C3-C50a, C50b/C50bp-C3-C3bm-C50f). Colony 9 showed the most distinct ITS2 type profile (C3k/C3bo-C50a-C3bp-C3) dominated by variant C3bo (38%), C3bp (~ 16%) and C3k (~ 15%). Additional sequencing of the rapidly evolving chloroplastic noncoding region marker psbAncr84 provided a finer delineation of inter- and intra-species variants within the Cladocopium genus. All psbAncr sequences belonged to the C3 radiation with colony 9 showing a distinct ASV profile from other colonies (Fig. 6b,c). ASVs from colony 1,11,12 were the most closely related to the recently formally described Cladocopium sodalum93 while ASVs from colony 9 were separated phylogenetically into another cluster presumably constituted of an undescribed C3 species.
Characterization of Symbiodiniaceae communities from 7 A.millepora colonies using ITS2 and psbAncr markers. (a) Relative abundance of ITS2 sequence variants assigned using Symportal analytical framework. The 30 most abundant variants are shown for plotting purposes and labels in bold indicate the 5 most abundant variants among all colonies. Predicted ITS2 profiles are plotted below (normalized to 1). (b) Relative abundance of the 20 most abundant psbAncr ASVs in DNA samples from colonies 1,9,11,12. (c) Maximum-likelihood phylogeny of the psbAncr ASVs and reference sequences from the C3-radiation (bootstrap support values based on 1000 replicates).
Effect of genotype on phenotypic response to heat stress
All colonies had comparable levels of genetic dissimilarity calculated on all SNPs, and no correlation was found between the phenotype dissimilarity matrix and the genotype dissimilarity matrix based on all SNPs (allelic dissimilarity; Mantel test p-value = 0.624). Conversely, by focusing on SNPs previously identified as candidate heat-associated loci (HAL), we found a significant correlation between genotype and phenotype dissimilarity matrices (Fig. 7a). The strength of the correlation increased when selecting SNPs significantly associated with bleaching alert frequency variables, being the highest for 22 SNPs with BAF association q-value < 0.03 (Mantel test, allelic dissimilarity: p.value = 0.029, euclidean distance: p.value = 0.005. When more stringent q-value thresholds were used (lower number of SNPs), the correlation decreased. Repeating this procedure using random SNPs instead yielded uniform p.value distributions at each sample size (as expected) and showed no effect of the sample size on the genotype x phenotype correlation (Fig. 7b, bottom panel).
Correlation between clustering genotypes based on candidate climate-adaptive loci and clustering of their phenotypic response to artificial heat stress assessed through Mantel tests between phenotype and genotype dissimilarity matrices. (a) For each point the genotype matrix is computed using Euclidean (blue) and allelic dissimilarity (red) distances on SNPs having an association q-value with one of the two bleaching alert variables (1 and 5 km) inferior to a specific threshold (X-axis). SNPs with q-values close to 0 display allelic frequencies strongly associated with thermal anomalies. The dashed line indicates the q-value threshold for which the best Mantel correlation p.value is obtained (q-value < 0.03, 22 SNPs). (b) The same procedure is repeated for genotype matrices with the same number of SNPs but randomly sampled among all SNPs. Boxplots display first quartile, median and third quartile from 1000 replications for each genotype matrix.
Top molecular markers associated with heat tolerance variation
The comparison of alleles between clusters revealed 4 SNPs among the top 22 distinguishing the colonies 10 and 11 from most of the others (e.g., homozygote reference in 10, 11 vs homozygote alternative in 1,3,9,12,13, Supplementary Tab. 6). The closest genes to these SNPs coded for an Oxysterol-binding protein-related protein 1 with a role in lipid binding and transport [GO:0006869] (SNP7), a cation-independent mannose-6-phosphate receptor involved in signaling activity [GO:0038023] and possibly apoptotic processes [GO:0043065] (SNP8) and a protein homolog to Golgi-associated plant pathogenesis-related protein 1 possibly involved in the MAP kinases ERK1/ERK2 pathways [GO:0070374] (SNP15). Of note, the closest coding sequence to the 4th SNPs did not match against any reference of the Uniprot/swissprot database. Additional SNPs separating colonies 12 and 13 from other clusters were found in proximity to genes coding for a Histone H2A (SNP4) and one protein containing Leucine-rich repeats and immunoglobulin-like domains (SNP11). Other SNPs that differed between colonies but did not differentiate clusters perfectly (Supplementary Tab. 6) were found in vicinity to genes coding for proteins such as the MICOS complex subunit MIC60, Interferon alpha-inducible protein 6, Low- density lipoprotein receptor related protein 6, Omega-scoloptoxin-Ssm1a and Enoyl- [acyl- carrier- protein] reductase.
Discussion
Candidate adaptive loci identified by seascape genomics correlate with differential response to artificial heat stress
Despite a growing number of candidate heat associated loci (HAL) identified by genotype environment association studies (GEAs) in scleractinian corals70,101,102,103, the relevance of these markers is rarely tested. Here, we used ex-situ, long-term heat stress experiment to investigate the link between genotypes at HAL revealed by seascape genomics and intrapopulation phenotypic variation in heat tolerance. We found that the clustering of colonies based on their alleles at candidate heat associated loci reflected the clustering based on phenotypic responses to artificial thermal stress. Heat tolerance is well known to be a polygenic trait in corals28,36,37,38,39,40,67,104, which is also supported by our results showing the best match between phenotypes and genotypes based on 22 HAL (represented by SNPs). Of note, this optimal number of loci is of the same order of magnitude (tens to hundreds) as reported in other studies28,31,38,105,106 despite discrepancies in genotyping methods.
Genes located in proximity of the SNPs that clearly discriminated tolerant and sensitive genotypes were associated with molecular functions known or supposed to be involved in heat stress response and cellular death except for one of unknown function. Some were associated with the MAP kinase pathway which acts on a wide variety of cell functions including cell survival and apoptosis and has been largely cited as a transduction pathway in coral bleaching107. Of interest, this pathway was also found as a candidate for adaptation in seascape genomics on Acropora digitifera in Japan102. One gene coded for a mannose-6-phosphate receptor involved in lysosomal biogenesis, a process known to have an important role in cellular homeostasis and possibly apoptotic processes108. Another gene possibly involved in the differences observed here, encodes for a histone H2A with a role in DNA binding and repair. Interestingly, H2A.X histone post-translational phosphorylation was shown to be impaired by nutrient and thermal stress in a previous study109 and could explain the hampering of DNA repair mechanisms and the negative effect of nutrients during bleaching. Finally, the results pointed out an Oxysterol-binding protein-related protein 1 which is known to be implicated in cholesterol trafficking between endosomes and endoplasmic reticulum110. Although the correlation remains purely speculative, formation of vesicles in the autophagic pathway has been suggested to play an interconnected role with apoptosis in cnidarian bleaching111.
Closely related symbiont species may alter holobiont response to heat stress
All colonies investigated in this experiment harbored Symbiodiniaceae belonging to the Cladocopium genus (C3-radiation). Although the short length of amplified sequence (~ 300 bp vs 800 bp) prevents a formal species identification, psbAncr phylogeny suggests that colonies were associated with Cladocopium sodalum93 or a closely related species. Interestingly colony 9 showed lower symbiont and chlorophyll retention than expected solely from the host genotype at the 22 HAL (phenotype clustering with colonies 1,3, Fig. 6; but genotype clustering with colonies 12,13, Supplementary Fig. 8), while hosting a distinct undescribed C3 species from other colonies. As important discrepancies in thermal tolerance can occur within Symbiodiniaceae genera112, the species harbored by this colony may be more sensitive to heat stress and exacerbate the holobiont health impairment. Of note, we only characterized the initial symbiont community composition upon collection in the field. Therefore, it is not possible to rule out a change in composition during the experiment that may have altered heat stress response.
Intrapopulation variation in heat tolerance
Observations of phenotypic responses to natural and artificial heat stress has revealed considerable intra-genus and intraspecific variation in heat tolerance among coral populations with distinct thermal histories48,113,114,115,116,117,118,119. Bleaching129 and mortality5,130 can be observed even under moderate heat stress (< 4 DHW) in sensitive taxa such as the Acropora genus. In particular, studies conducted on A. millepora reported an 82% decline in Symbiodiniaceae concentrations under 4.2 experimental DHW (eDHW131) and high bleaching under 4.5 eDHW74,132. Conversely, we found that 2 weeks at 2 °C above MMM (4 eDHW) did not lead to any significant paling or photosynthetic damage. This suggests higher average tolerance in populations frequently subjected to marine heat waves such as Snark reef (Supplementary Fig. 2) although other insidious negative effects may not have been captured by our physiological measurements (e.g., on growth and reproduction133).
Recent studies have also highlighted variation within populations48,120, with colonies sometimes able to tolerate > 4 Degree Heating Weeks (DHW) above thresholds of conspecific individuals from similar environments29. Here, we observed highly variable physiological responses both among replicate nubbins—possibly due to their position within the experimental tanks—and among individual colonies. For instance, average chlorophyll and symbiont density retention were > 80% higher under heat stress in the most tolerant colonies compared to the most sensitive ones. Increasing the sample size might reveal even higher level of intraspecific variation at this reef. For instance, Cunning et al.121 found that 7–8 colonies encompassed more than 50% of Florida A. cervicornis thermal variability, but larger sample sizes were required to have a high probability of capturing at least one heat tolerant genotype. Of interest, distinct response of physiological variables within tolerance groups suggests different mechanisms of tolerance and bleaching among colonies. For instance, despite comparable visual scores, colony 12 had lower symbiont cells density than colony 13 throughout heat stress, which may alleviate the production of reactive oxygen species48 as shown by its higher maintenance of photosynthetic performance. Colonies 10 and 11 both demonstrated low tolerance to heat stress but also differed in their physiological response. Colony 11 exhibited visible signs of bleaching, with depigmented polyps still visible in their calices (although less active) which eventually lead to the death of several nubbins on the long-term. Conversely, colony 10 experienced rapid tissue decay in a peeling form, with no visible polyps under a binocular magnifier which has been linked to bacterial and viral infections122,123,124 or extreme heat stress115,125. As it was neither observed for other colonies nor for colony 10 in the control treatment, it is unlikely to be the consequence of pathogen introduction through handling of the colony. However, high temperatures are known to promote microbial infections in many benthic organisms, including corals, notably through the disruption of host immune system functions126,127,128. As such, colony 10 might harbor a thermosensitive immune system, which could be a possible explanation for its health impairment in this experiment.
Challenges of building a tolerance index using SNPs data
Despite the identification of relevant molecular targets, building a tolerance index from SNP genotyping data that would predict individual thermal thresholds without prior information on phenotypes is highly challenging. First, many other factors than host and symbiont genotypes are susceptible to alter individual thermal thresholds. Factors such as epigenetics34, microbial communities134, microhabitats environmental conditions135 and seasonal fluctuations112 are for instance alternate hypotheses to explain the difference of clustering between colony 9 phenotype and genotype. Second, HAL used in this study have been identified across populations from New Caledonia but whether or not these markers have predictive power across other reef systems has not been tested yet. Third, interpreting results from GEAs is challenging as high incertitude remains on the genetic architecture of adaptation136. Corals heat tolerance is polygenic, thus driven by many loci of small effect size36, with possible epistasis (e.g., antagonist, synergistic, neutral) and functional redundancy between genomic regions and with other holobiont partners 61. The thermal tolerance benefit arising from having a certain allele at a HAL may therefore depend on allelic combination at many other loci as well as transcriptional and post-transcriptional regulation. Here, the limited sample size was unlikely to encompass all of the standing adaptive genetic diversity at this reef, as shown by the absence of clear colony allelic segregation at most of the 22 SNPs, and prevented testing the effect size of each individual loci. Limited sample size and uncertainty on loci effect sizes limits the predictive power of polygenic scores at the genotype level (Supplementary Fig. 4). Additional loci involved in thermal adaptation may also have not been captured by seascape genomics, for instance if local adaptation occurs in ways contradicting the method underlying hypothesis136. Finally, the sequencing technique used here is not well-suited for the identification of adaptive signals in non-coding regions, and some of the loci previously identified were discarded in this analysis as they were not genotyped in all colonies. Increasing affordability of deep-sequencing and whole-genome sequencing techniques (e.g., see137) will help to overcome these representation barriers.
Combining sets of genes from different methods and on different life stages (adults, larvae, juveniles) can help to strengthen our confidence in adaptive role of specific loci. For instance, Jin et al.138 used population genetics and gene by environment analysis to identify a restricted set of genes possibly involved in heat tolerance that was further refined using a natural bleaching experiment and gene by phenotype analysis. Ultimately, association studies are correlative methods thus full validation of HAL will require approaches such as functional genomics and reverse genetics. As such, the recent development of CRISPR/Cas-9-mediated genome editing in corals139,140 can be exploited in controlled experiments to validate the effect of genetic markers on heat tolerance and better understand the role of orphan genes141.
Conclusion
In this study we showed that candidate heat adaptive loci (HAL) revealed by seascape genomics correlate with phenotypic response from conspecific coral colonies to artificial heat stress. These results add to the growing body of evidence highlighting the importance of harnessing adaptive genetic variation to foster coral reef resilience58. For instance, genetic markers can be used to target tolerant individuals for sexual propagation, selective breeding and assisted gene flow or prioritize conservation on reefs with high proportion of heat adapted alleles46,59,60,142,143. Due to the limited sample size, we were not able to evaluate the predictive accuracy of these HAL. Their integration in restoration and conservation frameworks therefore requires further validation, for instance by recording bleaching phenotypes during natural marine heatwaves across a larger number of individuals of known genotypes, or through the development of reverse genetics in corals.
Data availability
Raw data of DaRT sequencing are available on BioProject, no. PRJNA888450 (NCBI), raw data of ITS2 metarbacoding of Symbiodiniaceae on BioProject, no PRJNA888797 (NCBI) and raw data of psbAncr on BioProject, no PRJNA949731 (NCBI).
References
Hoegh-Guldberg, O. et al. Impacts of 1.5 C global warming on natural and human systems. Global warming of 1.5 C. An IPCC Special Report (2018).
De’ath, G., Fabricius, K. E., Sweatman, H. & Puotinen, M. The 27–year decline of coral cover on the great barrier reef and its causes. Proc. Natl. Acad. Sci. 109, 17995–17999 (2012).
Gardner, T. A., Côté, I. M., Gill, J. A., Grant, A. & Watkinson, A. R. Long-term region-wide declines in caribbean corals. Science 301, 958–960 (2003).
Bruno, J. F. & Selig, E. R. Regional decline of coral cover in the Indo-Pacific: Timing, extent, and subregional comparisons. PLoS ONE 2, e711 (2007).
Hughes, T. P. et al. Global warming and recurrent mass bleaching of corals. Nature 543, 373–377 (2017).
Eddy, T. D. et al. Global decline in capacity of coral reefs to provide ecosystem services. One Earth 4, 1278–1285 (2021).
Weis, V. M. Cellular mechanisms of cnidarian bleaching: Stress causes the collapse of symbiosis. J. Exp. Biol. 211, 3059–3066 (2008).
Roberty, S. & Plumier, J. C. Bleaching physiology: Who’s the ‘weakest link’—Host vs. symbiont?. Emerg. Top. Life Sci. 6, 17–32 (2022).
Hoegh-Guldberg, O. Climate change, coral bleaching and the future of the world’s coral reefs. Mar. Freshw. Res. 50, 839–866 (1999).
Oliver, E. C. et al. Longer and more frequent marine heatwaves over the past century. Nat. commun. 9, 1–12 (2018).
Hughes, T. Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359, 80–83 (2018).
Van Hooidonk, R. et al. Local-scale projections of coral reef futures and implications of the Paris agreement. Sci. Rep. 6, 1–8 (2016).
IPCC, Summary for Policymakers. In: Global Warming of 1.5°C. An IPCC Special Report on the impacts of global warming of 1.5°C above pre-industrial levels and related global greenhouse gas emission pathways, in the context of strengthening the global response to the threat of climate change, sustainable development, and efforts to eradicate poverty (eds. Masson-Delmotte, V. et al). Cambridge University Press, Cambridge, USA, pp. 3–24, https://doi.org/10.1017/9781009157940.001 (2018).
McClanahan, T. R. et al. Large geographic variability in the resistance of corals to thermal stress. Glob. Ecol, Biogeogr. 29, 2229–2247. https://doi.org/10.1111/geb.13191 (2020).
Muir, P. R., Done, T. & Aguirre, D. J. High regional and intra-generic variation in susceptibility to mass bleaching in Indo-Pacific coral species. Glob. Ecol. Biogeogr. 30, 1889–1898 (2021).
Raja, N. B. et al. Morphological traits of reef corals predict extinction risk but not conservation status. Glob. Ecol. Biogeogr. 30(8), 1597–1608 (2021).
Schoepf, V., Sanderson, H. & Larcombe, E. Coral heat tolerance under variable temperatures: Effects of different variability regimes and past environmental history vs. current exposure. Limnol. Oceanogr. 67, 404–418 (2021).
Page, C. E. et al. Seeking resistance in coral reef ecosystems: The interplay of biophysical factors and bleaching resistance under a changing climate: The interplay of a reef’s biophysical factors can mitigate the coral bleaching response. BioEssays 41, 1800226 (2019).
Wiedenmann, J. et al. Nutrient enrichment can increase the susceptibility of reef corals to bleaching. Nat. Clim. Ch. 3, 160–164 (2013).
Burkepile, D. E. et al. Nitrogen identity drives differential impacts of nutrients on coral bleaching and mortality. Ecosystems 23, 798–811 (2020).
Gonzalez-Espinosa, P. C. & Donner, S. D. Cloudiness reduces the bleaching response of coral reefs exposed to heat stress. Glob. Ch. Biol. 27(15), 3474–3486 (2021).
van Oppen, M. J., Baker, A. C., Coffroth, M. A. & Willis, B. L. Bleaching Resistance and the Role of Algal Endosymbionts in Coral bleaching (Springer, 2009).
Levin, R. A. et al. Sex, scavengers, and chaperones: Transcriptome secrets of divergent symbiodinium thermal tolerances. Mol. Biol. Evol. 33, 2201–2215 (2016).
van Oppen, M. J. & Blackall, L. L. Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 17, 557–567 (2019).
Torda, G. et al. Rapid adaptive responses to climate change in corals. Nat. Clim. Ch. 7, 627–636 (2017).
Cunning, R., Ritson-Williams, R. & Gates, R. D. Patterns of bleaching and recovery of Montipora capitata in Kāne ‘ohe bay, Hawai ‘i, USA. Mar. Ecol. Prog. Ser. 551, 131–139 (2016).
Voolstra, C. R. et al. Standardized short-term acute heat stress assays resolve historical differences in coral thermotolerance across microhabitat reef sites. Glob. Ch. Biol. 26, 4328–4343 (2020).
Drury, C. et al. Intrapopulation adaptive variance supports thermal tolerance in a reef-building coral. Commun. Biol. 5, 1–10 (2022).
Humanes, A. et al. Within-population variability in coral heat tolerance indicates climate adaptation potential. Proc. R. Soc. B 289, 20220872 (2022).
Bay, R. A. & Palumbi, S. R. Rapid acclimation ability mediated by transcriptome changes in reef-building corals. Genome Biol. Evol. 7, 1602–1612 (2015).
Barshis, D. J. et al. Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. 110, 1387–1392 (2013).
Camp, E. F. et al. Corals exhibit distinct patterns of microbial reorganisation to thrive in an extreme inshore environment. Coral Reefs 39(3), 701–716 (2020).
Berkelmans, R. & Van Oppen, M. J. The role of zooxanthellae in the thermal tolerance of corals: A ‘nugget of hope’for coral reefs in an era of climate change. Proc. R. Soc. B Biol. Sci. 273, 2305–2312 (2006).
Liew, Y. J. et al. Intergenerational epigenetic inheritance in reef-building corals. Nat. Clim. Ch. 10, 254–259 (2020).
Lesser, M., Stat, M. & Gates, R. The endosymbiotic dinoflagellates (Symbiodinium sp.) of corals are parasites and mutualists. Coral Reefs 32, 603–611 (2013).
Rose, N. H., Bay, R. A., Morikawa, M. K. & Palumbi, S. R. Polygenic evolution drives species divergence and climate adaptation in corals. Evolution 72, 82–94 (2018).
Dixon, G. B. et al. Genomic determinants of coral heat tolerance across latitudes. Science 348, 1460–1462 (2015).
Kirk, N. L., Howells, E. J., Abrego, D., Burt, J. A. & Meyer, E. Genomic and transcriptomic signals of thermal tolerance in heat-tolerant corals (Platygyra daedalea) of the Arabian/Persian gulf. Mol. Ecol. 27, 5180–5194 (2018).
Smith, E. G. et al. Signatures of selection underpinning rapid coral adaptation to the world warmest reefs. Sci. Adv. 8, eabl7287 (2022).
Rose, N. H. et al. Genomic analysis of distinct bleaching tolerances among cryptic coral species. Proc. R. Soc. B 288, 20210678 (2021).
Sully, S., Burkepile, D. E., Donovan, M., Hodgson, G. & Van Woesik, R. A global analysis of coral bleaching over the past two decades. Nat. Commun. 10, 1–5 (2019).
DeCarlo, T. M. et al. Acclimatization of massive reef-building corals to consecutive heatwaves. Proc. R. Soc. B 286, 20190235 (2019).
Shlesinger, T. & van Woesik, R. Oceanic differences in coral-bleaching responses to marine heatwaves. Sci. Total Environ. 871, 162113 (2023).
Bairos-Novak, K. A. O., Hoogenboom, M. A. O., van Oppen, M. A. O. & Connolly, S. A. O. Coral adaptation to climate change: Meta-analysis reveals high heritability across multiple traits. Glob. Ch. Biol. 27, 5694–5710 (2021).
Elder, H. et al. Genetic variation in heat tolerance of the coral Platygyra daedalea indicates potential for adaptation to ocean warming. BioRxiv 27(22), 5694 (2020).
Howells, E. J. et al. Enhancing the heat tolerance of reef-building corals to future warming. Sci. Adv. 7, eabg6070 (2021).
Wright, R. M. et al. Positive genetic associations among fitness traits support evolvability of a reef-building coral under multiple stressors. Glob. Ch. Biol. 25, 3294–3304 (2019).
Cornwell, B. et al. Widespread variation in heat tolerance and symbiont load are associated with growth tradeoffs in the coral Acropora hyacinthus in Palau. Elife 10, e64790 (2021).
Jones, A. M. & Berkelmans, R. Tradeoffs to thermal acclimation: Energetics and reproduction of a reef coral with heat tolerant symbiodinium type-D. J. Mar. Biol. 2011, 1–12 (2011).
Bay, L. K. & Howells, E. J. Ecosystem resilience: Mapping the future for coral reefs. eLife 10, e72978 (2021).
Bay, R. A., Rose, N. H., Logan, C. A. & Palumbi, S. R. Genomic models predict successful coral adaptation if future ocean warming rates are reduced. Sci. Adv. 3, e1701413 (2017).
Quigley, K. M., Bay, L. K. & van Oppen, M. J. The active spread of adaptive variation for reef resilience. Ecol. Evol. 9, 11122–11135 (2019).
Logan, C. A., Dunne, J. P., Ryan, J. S., Baskett, M. L. & Donner, S. D. Quantifying global potential for coral evolutionary response to climate change. Nat. Clim. Ch. 11(6), 537–542 (2021).
Matz, M. V., Treml, E. A. & Haller, B. C. Estimating the potential for coral adaptation to global warming across the Indo-West Pacific. Glob. Ch. Biol. 26, 3473–3481 (2020).
Mumby, P. J., Mason, R. A. & Hock, K. Reconnecting reef recovery in a world of coral bleaching. Limnol. Oceanogr. Methods 19(10), 702–713 (2021).
Ainsworth, T. D. et al. Rebuilding relationships on coral reefs: Coral bleaching knowledge‐sharing to aid adaptation planning for reef users: Bleaching emergence on reefs demonstrates the need to consider reef scale and accessibility when preparing for, and responding to, coral bleaching. BioEssays 43, 2100048 (2021).
Colton, M. A. et al. Coral conservation in a warming world must harness evolutionary adaptation. Nat. Ecol. Evol. 6, 1405–1407 (2022).
Howells, E.J., Bay, L.K., Bay, R.A. Identifying, Monitoring, and Managing Adaptive Genetic Variation in Reef-Building Corals under Rapid Climate Warming. In: (eds. van Oppen, M. J. H., Aranda Lastra, M.) Coral Reef Conservation and Restoration in the Omics Age. Coral Reefs of the World. 15, 55–70. https://doi.org/10.1007/978-3-031-07055-6_4 (2022).
Humanes, A. et al. An experimental framework for selectively breeding corals for assisted evolution. Front. Mar. Sci. 8, 626 (2021).
van Oppen, M. J., Oliver, J. K., Putnam, H. M. & Gates, R. D. Building coral reef resilience through assisted evolution. Proc. Natl. Acad. Sci. 112, 2307–2313 (2015).
Avila-Magaña, V. et al. Elucidating gene expression adaptation of phylogenetically divergent coral holobionts under heat stress. Nat. Commun. 12, 1–16 (2021).
DeSalvo, M. et al. Differential gene expression during thermal stress and bleaching in the caribbean coral Montastraea faveolata. Mol. Ecol. 17, 3952–3971 (2008).
Thomas, L. et al. Mechanisms of ecological divergence with gene flow in a reef-building coral on an isolated atoll in Western Australia. BioRxiv 5, 1 (2021).
Tam, V. et al. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 20, 467–484 (2019).
Pharoah, P. D., Dunning, A. M., Ponder, B. A. & Easton, D. F. Association studies for finding cancer-susceptibility genetic variants. Nat. Rev. Cancer 4, 850–860 (2004).
Hirschhorn, J. N. & Daly, M. J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6, 95–108 (2005).
Fuller, Z. L. et al. Population genetics of the coral Acropora millepora: Toward genomic prediction of bleaching. Science https://doi.org/10.1126/science.aba4674 (2020).
van Woesik, R. et al. Coral-bleaching responses to climate change across biological scales. Glob. Ch. Biol. 28(14), 4229–4250 (2022).
Riginos, C., Crandall, E. D., Liggins, L., Bongaerts, P. & Treml, E. A. Navigating the currents of seascape genomics: How spatial analyses can augment population genomic studies. Curr. Zool. 62, 581–601 (2016).
Selmoni, O. et al. Seascape genomics reveals candidate molecular targets of heat stress adaptation in three coral species. Mol. Ecol. 30, 1892–1906 (2021).
Loya, Y. et al. Coral bleaching: The winners and the losers. Ecol. Lett. 4, 122–131 (2001).
Wallace, C. C., Done, B. J. & Muir, P. R. Revision and catalogue of worldwide staghorn corals Acropora and Isopora (Scleractinia: Acroporidae) in the museum of tropical Queensland. MQMN 57, 1–255 (2012).
Ying, H. et al. The whole-genome sequence of the coral Acropora millepora. Genome Biol. Evol. 11, 1374–1379 (2019).
Lutz, A., Raina, J. B., Motti, C. A., Miller, D. J. & van Oppen, M. J. Host coenzyme Q redox state is an early biomarker of thermal stress in the coral Acropora millepora. PLoS ONE 10, e0139290 (2015).
Pernice, M. et al. Regulation of apoptotic mediators reveals dynamic responses to thermal stress in the reef building coral Acropora millepora. PLoS ONE 6, e16095 (2011).
Császár, N., Seneca, F. & Van Oppen, M. Variation in antioxidant gene expression in the scleractinian coral Acropora millepora under laboratory thermal stress. Mar. Ecol. Prog. Ser. 392, 93–102 (2009).
Grottoli, A. G. et al. Increasing comparability among coral bleaching experiments. Ecol. Appl. 31, e02262 (2021).
Santoro, E. P. et al. Coral microbiome manipulation elicits metabolic and genetic restructuring to mitigate heat stress and evade mortality. Sci. Adv. 7, eabg3088 (2021).
Ferrier-Pagès, C. et al. Summer autotrophy and winter heterotrophy in the temperate symbiotic coral Cladocora caespitosa. Limnol. Oceanogr. 56, 1429–1438 (2011).
Jeffrey, S. T. & Humphrey, G. New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem. Physiol. Pflanzen 167, 191–194 (1975).
Naumann, M. S., Niggl, W., Laforsch, C., Glaser, C. & Wild, C. Coral surface area quantification–evaluation of established techniques by comparison with computer tomography. Coral Reefs 28, 109–117 (2009).
Rouzé, H. et al. An updated assessment of Symbiodinium spp. that associate with common scleractinian corals from Moorea (French Polynesia) reveals high diversity among background symbionts and a novel finding of clade B. PeerJ 5, e2856 (2017).
Kilian, A. et al. Data Production and Analysis in Population Genomics (Springer, 2012).
Moore, R. B., Ferguson, K. M., Loh, W. K., Hoegh-Guldberg, O. & Carter, D. A. Highly organized structure in the non-coding region of the psbA minicircle from clade C symbiodinium. Int. J. Syst. Evolut. Microbiol. 53, 1725–1734 (2003).
Davies, S. et al. Building consensus around the assessment and interpretation of Symbiodiniaceae diversity. PeerJ 11, e15023 (2023).
Hume, B. C. et al. An improved primer set and amplification protocol with increased specificity and sensitivity targeting the symbiodinium ITS2 region. PeerJ 6, e4816 (2018).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Hume, B. C. et al. SymPortal: A novel analytical framework and platform for coral algal symbiont next-generation sequencing ITS2 profiling. Mol. Ecol. Resour. 19, 1063–1080 (2019).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Callahan, B. J., McMurdie, P. J. & Holmes, S. P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643 (2017).
Sayers, E. W. et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 49, D10 (2021).
McMurdie, P. J. & Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 (2013).
Butler, C. C. et al. Formal recognition of host-generalist species of dinoflagellate (Cladocopium, Symbiodiniaceae) mutualistic with Indo-Pacific reef corals. J. Phycol. 59(4), 698–711 (2023).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547 (2018).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ (2021).
Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Soft. https://doi.org/10.18637/jss.v067.i01 (2014).
Lenth R. emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 1.10.2. https://CRAN.R-project.org/package=emmeans (2024).
Liu, G., Strong, A. E. & Skirving, W. Remote sensing of sea surface temperatures during 2002 barrier reef coral bleaching. Eos Trans. Am. Geophys. Un. 84, 137–141 (2003).
Suzuki, R. & Shimodaira, H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 (2006).
Wickham, H. ggplot2. WIREs Comput. Stat. 3, 180–185 (2011).
Lundgren, P., Vera, J. C., Peplow, L., Manel, S. & van Oppen, M. J. Genotype–environment correlations in corals from the great barrier reef. BMC Genet. 14, 1–16 (2013).
Selmoni, O., Rochat, E., Lecellier, G., Berteaux-Lecellier, V. & Joost, S. Seascape genomics as a new tool to empower coral reef conservation strategies: An example on north-western Pacific Acropora digitifera. Evolut. Appl. 13, 1923–1938 (2020).
Adam, A. A., Thomas, L., Underwood, J., Gilmour, J. & Richards, Z. T. Population connectivity and genetic offset in the spawning coral Acropora digitifera in Western Australia. Mol. Ecol. 31(13), 3533–3547 (2022).
Cooke, I. et al. Genomic signatures in the coral holobiont reveal host adaptations driven by holocene climate change and reef specific symbionts. Sci. Adv. 6, eabc6318. https://doi.org/10.1126/sciadv.abc6318 (2020).
Bay, R. A. & Palumbi, S. R. Multilocus adaptation associated with heat resistance in reef-building corals. Curr. Biol. 24, 2952–2956 (2014).
Quigley, K. M., Bay, L. K. & van Oppen, M. J. Genome-wide SNP analysis reveals an increase in adaptive genetic variation through selective breeding of coral. Mol. Ecol. 29, 2176–2188 (2020).
Szabó, M., Larkum, A. W. & Vass, I. A Review: The Role of Reactive Oxygen Species in Mass Coral Bleaching (Cham, 2020).
Siva Kumar, N. & Bhamidimarri, M. P. Lysosomal enzymes and their receptors in invertebrates: An evolutionary perspective. Curr. Protein Pept. Sci. 16, 49–65 (2015).
Rodriguez-Casariego, J. A. et al. Coral epigenetic responses to nutrient stress: Histone H2A. X phosphorylation dynamics and DNA methylation in the staghorn coral Acropora cervicornis. Ecol. Evol. 8, 12193–12207 (2018).
Zhao, K., Foster, J. & Ridgway, N. D. Oxysterol-binding protein-related protein 1 variants have opposing cholesterol transport activities from the endolysosomes. Mol. Biol. Cell 31, 793–802 (2020).
Dunn, S. R., Schnitzler, C. E. & Weis, V. M. Apoptosis and autophagy as mechanisms of dinoflagellate symbiont release during cnidarian bleaching: Every which way you lose. Proc. R. Soc. B Biol. Sci. 274, 3079–3085 (2007).
Sampayo, E., Ridgway, T., Bongaerts, P. & Hoegh-Guldberg, O. Bleaching susceptibility and mortality of corals are determined by fine-scale differences in symbiont type. Proc. Natl. Acad. Sci. 105, 10444–10449 (2008).
Thomas, L. et al. Mechanisms of thermal tolerance in reef-building corals across a fine-grained environmental mosaic: Lessons from Ofu American Samoa. Front. Mar. Sci. 4, 434 (2018).
Howells, E. J., Berkelmans, R., van Oppen, M. J., Willis, B. L. & Bay, L. K. Historical thermal regimes define limits to coral acclimatization. Ecology 94, 1078–1088 (2013).
Evensen, N. R., Fine, M., Perna, G., Voolstra, C. R. & Barshis, D. J. Remarkably high and consistent tolerance of a red sea coral to acute and chronic thermal stress exposures. Limnol. Oceanogr. 66(5), 1718–1729 (2021).
Evensen, N. R. et al. Empirically derived thermal thresholds of four coral species along the red sea using a portable and standardized experimental approach. Coral Reefs 41(2), 239–252 (2022).
Klepac, C. N. & Barshis, D. J. High-resolution in situ thermal metrics coupled with acute heat stress experiments reveal differential coral bleaching susceptibility. Coral Reefs 41(4), 1045–1057 (2022).
Marzonie, M. R. et al. The effects of marine heatwaves on acute heat tolerance in corals. Glob. Ch. Biol. 29(2), 404–416 (2022).
Voolstra, C. R. et al. Contrasting heat stress response patterns of coral holobionts across the red sea suggest distinct mechanisms of thermal tolerance. Mol. Ecol. 30, 4466–4480 (2021).
Walker, N. S., Nestor, V., Golbuu, Y. & Palumbi, S. R. Coral bleaching resistance variation is linked to differential mortality and skeletal growth during recovery. Evolut. Appl. 16(2), 504–517 (2022).
Cunning, R. et al. Census of heat tolerance among Florida’s threatened staghorn corals finds resilient individuals throughout existing nursery populations. Proc. R. Soc. B 288, 20211613 (2021).
Lawrence, S. A., Davy, J. E., Aeby, G. S., Wilson, W. H. & Davy, S. K. Quantification of virus-like particles suggests viral infection in corals affected by porites tissue loss. Coral Reefs 33, 687–691 (2014).
Meyer, J. L. et al. Microbial community shifts associated with the ongoing stony coral tissue loss disease outbreak on the florida reef tract. Front. Microbiol. 10, 2244 (2019).
Luna, G., Biavasco, F. & Danovaro, R. Bacteria associated with the rapid tissue necrosis of stony corals. Environ. Microbiol. 9, 1851–1857 (2007).
Leggat, W. P. et al. Rapid coral decay is associated with marine heatwave mortality events on reefs. Curr. Biol. 29, 2723–2730 (2019).
Ambalavanan, L., Iehata, S., Fletcher, R., Stevens, E. H. & Zainathan, S. C. A review of marine viruses in coral ecosystem. J. Mar. Sci. Eng. 9, 711 (2021).
McDevitt-Irwin, J. M., Baum, J. K., Garren, M. & Vega Thurber, R. L. Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 4, 262 (2017).
van de Water, J. A. et al. Antimicrobial and stress responses to increased temperature and bacterial pathogen challenge in the holobiont of a reef-building coral. Mol. Ecol. 27, 1065–1080 (2018).
Sakai, K., Singh, T. & Iguchi, A. Bleaching and post-bleaching mortality of acropora corals on a heat-susceptible reef in 2016. PeerJ 7, e8138 (2019).
Hughes, T. P. et al. Global warming transforms coral reef assemblages. Nature 556, 492–496 (2018).
Leggat, W., Heron, S. F., Fordyce, A., Suggett, D. J. & Ainsworth, T. D. Experiment degree heating week (eDHW) as a novel metric to reconcile and validate past and future global coral bleaching studies. J. Environ. Manag. 301, 113919 (2022).
Bellantuono, A. J., Hoegh-Guldberg, O. & Rodriguez-Lanetty, M. Resistance to thermal stress in corals without changes in symbiont composition. Proc. R. Soc. B Biol. Sci. 279, 1100–1107 (2012).
Leinbach, S. E., Speare, K. E., Rossin, A. M., Holstein, D. M. & Strader, M. E. Energetic and reproductive costs of coral recovery in divergent bleaching responses. Sci. Rep. 11, 1–10 (2021).
Voolstra, C. R. et al. Extending the natural adaptive capacity of coral holobionts. Nat. Rev. Earth Environ. https://doi.org/10.1038/s43017-021-00214-3 (2021).
Brown, K. T., Eyal, G., Dove, S. G. & Barott, K. L. Fine-scale heterogeneity reveals disproportionate thermal stress and coral mortality in thermally variable reef habitats during a marine heatwave. Coral Reefs 42(1), 131–142 (2022).
Lasky, J. R., Josephs, E. B. & Morris, G. P. Genotype–environment associations to reveal the molecular basis of environmental adaptation. Plant Cell 35, 125–138 (2023).
Yoshioka, Y. et al. Whole-genome transcriptome analyses of native symbionts reveal host coral genomic novelties for establishing coral-algae symbioses. Genome Biol. Evol. 13, evaa240 (2021).
Jin, Y. K. et al. Genetic markers for antioxidant capacity in a reef-building coral. Sci. Adv. 2, e1500842 (2016).
Cleves, P. A., Strader, M. E., Bay, L. K., Pringle, J. R. & Matz, M. V. CRISPR/Cas9-mediated genome editing in a reef-building coral. Proc. Natl. Acad. Sci. USA 115, 5235–5240. https://doi.org/10.1073/pnas.1722151115 (2018).
Cleves, P. A. et al. Reduced thermal tolerance in a coral carrying CRISPR-induced mutations in the gene for a heat-shock transcription factor. Proc. Natl. Acad. Sci. 117, 28899–28905 (2020).
Cleves, P. A., Shumaker, A., Lee, J., Putnam, H. M. & Bhattacharya, D. Unknown to known: Advancing knowledge of coral gene function. Trends Genetics 36, 93–104 (2020).
Caruso, C., Hughes, K. & Drury, C. Selecting heat-tolerant corals for proactive reef restoration. Front. Mar. Sci. 8, 584 (2021).
Van Oppen, M. J. et al. Shifting paradigms in restoration of the world’s coral reefs. Glob. Ch. Biol. 23, 3437–3448 (2017).
Acknowledgements
We thank Olivier Chateau and the Aquarium des Lagons for their collaboration on running ex-situ aquarium experiments. We thank Pascal Dumas for providing control equipment for the experimental system. We are thankful to Reef Shelter for hosting the colonies in their coral garden. We also thank Clarisse Majorel, Christine Sidobre, Christophe Peignon and Mahe Dumas for their help with the collection and fragmentation of colonies. Finally, we thank Antoine Briand and Claire Boitel for their help with photosynthetic measurements.
Author information
Authors and Affiliations
Contributions
H.D., O.S., G.L. and V.B.L. initiated the project and designed the experiment. H.D. and H.G. built the experimental system. H.D., G.L. and V.B.L. collected and fragmented the colonies and recorded physiological parameters throughout the experiment along with T.J. H.D performed DNA extraction, chlorophyll and symbiont density measurements. H.D. performed the data analysis, supported by O.S., C.C.B., G.L. and V.B.L. H.D. wrote the first version of the manuscript, which was critically revised by all other authors. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Denis, H., Selmoni, O., Gossuin, H. et al. Climate adaptive loci revealed by seascape genomics correlate with phenotypic variation in heat tolerance of the coral Acropora millepora. Sci Rep 14, 22179 (2024). https://doi.org/10.1038/s41598-024-67971-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-67971-1