Abstract
Rabies virus (RABV) is the causative agent of rabies, a lethal neurological disease in mammals. RABV strains can be classified into fixed strains (laboratory strains) and street strains (field/clinical strains), which have different properties including cell tropism and neuroinvasiveness. RABV Toyohashi strain is a street strain isolated in Japan from an imported case which had been bitten by rabid dog in the Philippines. In order to facilitate molecular studies of RABV, we established a reverse genetics (RG) system for the study of the Toyohashi strain. The recombinant virus was obtained from a cDNA clone of Toyohashi strain and exhibited similar growth efficiency as the original virus in cultured cell lines. Both the original and recombinant strains showed similar pathogenicity with high neuroinvasiveness in mice, and the infected mice developed a long and inconsistent incubation period, which is characteristic of street strains. We also generated a recombinant Toyohashi strain expressing viral phosphoprotein (P protein) fused with the fluorescent protein mCherry, and tracked the intracellular dynamics of the viral P protein using live-cell imaging. The presented reverse genetics system for Toyohashi strain will be a useful tool to explore the fundamental molecular mechanisms of the replication of RABV street strains.
Similar content being viewed by others
Introduction
Rabies virus (RABV), a member of the genus Lyssavirus of the family Rhabdoviridae, is the causative agent of rabies, a lethal neurological disease in mammals. Due to the lack of an effective treatment and insufficient use of current post-exposure prophylaxis, it is estimated that rabies causes 13,000−60,000 human death annually, primarily in developing countries in Asia and Africa1,2,3. Therefore, the development of treatments and better prophylactic agents is desirable, and to this end further accumulation of basic knowledge at the molecular level on the infection, proliferation and pathogenesis of RABV is essential.
RABV strains consists of two groups: street strains, which are field strains isolated from a human patient or an animal with rabies, and fixed strains, which include laboratory strains and vaccine strains established through multiple passages of street strains in cultured cells and/or animals. Compared with fixed strains, street strains have different characteristics, such as more variable and longer incubation period and higher pathogenicity after peripheral infections (neuroinvasiveness)4,5. However, studies on the molecular mechanisms of RABV pathogenesis have mainly been carried out by using fixed strains, and little is known about the pathogenic mechanisms of street strains6,7,8. The RABV street strain Toyohashi was originally isolated in Japan from an imported rabies case infected with RABV in the Philippines and was shown to phylogenetically cluster with RABVs from dogs in the Phillipines9. In the Philippines, rabies is a significant public health problem, causing more than 300 human fatal cases each year10. Given the distinct genetic diversity of RABV, characterization of Toyohashi strain as the currently prevalent RABV strain in the Philippines will help us to better understand the pathogenesis of rabies.
A reverse genetics (RG) system allows us to manipulate the viral genome and obtain recombinant RABVs carrying amino acid substitutions or reporter gene expressing cassettes. To date, several street strains, including Komatsugawa (from a dog in Japan), 1088 (from a woodchuck in USA), 8743THA (from a human in Thailand), RABV-Fox (from a fox in Germany), RABV-Dog (from a dog in Azerbaijan), QS-05 (from a dog in Thailand), GD-SH-01 (from a pig in China) and SHBRV-18 (from a silver-haired bat in USA) have been successfully recovered by RG system, and used for research on their genetic and virological properties11,12,13,14,15,16,17,18,19,20. Recent large-scale phylogenetic analysis of RABV revealed that the Asian clade can be subdivided into 5 subclades from South-East Asia (SEA) 1–521,22. According to this classification, Philippine endemic strains belong to the subclade SEA 4, which has not yet been detected in any other countries. Collectively, the Toyohashi strain represents a different evolutionary lineage from the street strains previously applied to RG system for RABV9,22.
In this study, we established an RG system for the Toyohashi strain. The recombinant Toyohashi strain was recovered from cloned cDNA and had the same virological characteristics as the original virus as a street strain from the aspects of both in vitro and in vivo. Furthermore, we generated a recombinant Toyohashi strain encoding the viral phosphoprotein (P protein) fused with the fluorescent protein mCherry and applied this to live-cell imaging for tracking the intracellular movement of the viral P protein.
Results
In vitro growth kinetics of recombinant RABV Toyohashi strain (rToyo) recovered from a cDNA clone
In this study, we used RABV Toyohashi P2I-2M strain (Toyohashi), which was obtained from a human specimen by passaging twice in the brains of suckling mice and two times in NA cells9. To establish an RG system for the Toyohashi strain, the cDNA of full-length RABV genome was obtained from viral RNA of Toyohashi by RT-PCR and inserted into a pUC19 cloning site flanked by a T7 promoter sequence and a hepatitis delta virus (HDV) antigenomic ribozyme cDNA (Fig. 1A). Recombinant infectious RABV Toyohashi strain (rToyo) was successfully rescued from BHK/T7-9 cells transfected with the full-length genome plasmid and helper plasmids as described in the Materials and Methods section.
Growth efficiency of the recombinant RABV Toyohashi strain (rToyo) in cell cultures. (A) Schematic representation of the plasmid construct encoding the wild-type Toyohashi strain (Toyohashi, GenBank LC812291, 11,922 nt in size). The full genome cDNA is flanked by the T7 promoter sequence and the antigenomic hepatitis delta virus ribozyme (HDV-Rbz) in a pUC19 vector. (B) Mouse neuroblastoma (NA) cells and mouse myoblasts (G-8) were inoculated with Toyohashi (blue square) or rToyo (red circle) at a multiplicity of infection (MOI) of 0.01. The culture supernatants containing progeny viruses were collected at the indicated time points and titrated using a focus forming assay. Means ± standard deviations of triplicate data are shown in the graph. Statistics were calculated by a multiple t-test with Welch correction. *p < 0.05; ns, not significant (p ≧ 0.05).
We examined the growth kinetics of rToyo in mouse neuronal NA cells and mouse myoblast G-8 cells. Multiple-step growth curves of rToyo and Toyohashi were determined based on virus titers in the culture supernatants at different time points. The titers of both rToyo and Toyohashi increased exponentially in the cell lines, with maximum titers of 107 FFU/ml in NA cells, and 104 FFU/ml in G-8 cells (Fig. 1B). There was no statistically significant difference between titers of rToyo and Toyohashi at each day post infection (dpi) in either cells.
Pathogenicity of rToyo in mice
We next compared the virulence of rToyo with Toyohashi and other RABV strains using a mouse model. RABV street strain Yokohama, fixed strains challenge virus standard (CVS) and recombinant CVS (rCVS) were included as comparator strains in these experiments. Mice intramuscularly (i.m.) inoculated with these RABV strains developed body weight loss, piloerection, hindlimb paralysis approximately two days before reaching the humane endpoint. The median lethal dose (LD50) value of rToyo (102.0 FFU) was comparable to those of both street strains Toyohashi (101.7 FFU) and Yokohama (102.3 FFU), and was at least 100-fold lower than the LD50 values of fixed strain CVS (104.8 FFU) (Table 1). The disease progression of mice inoculated with street strains were variable (Fig. 2A), whereas most mice inoculated with CVS or rCVS developed severe body weight loss and reached the endpoint criteria within 8 dpi (Fig. 2B). There was no significant difference in survival rates between the mice inoculated at a dose of 102 FFU of rToyo, Toyohashi and the Yokohama strains (Fig. 2A). The incubation period was defined as the period from the day of viral infection until the weight of the inoculated mice decreased by more than 5% of their maximum body weight at the previous day (Supplementary Fig. S1). Notably, the incubation period of mice infected with rToyo was variable ranging from 7 to 17 days and was similar to those of Toyohashi and the Yokohama strain, while the incubation periods of mice infected with CVS and rCVS strains were 5–6 days which is shorter than that with rToyo (Fig. 2C). These results indicated that the recombinant and original Toyohashi strains had higher neuroinvasiveness and produced a longer and more inconsistent incubation periods compared to the CVS strain.
Pathogenicity of rToyo following intramuscular inoculation. Six-week-old male ddY mice were inoculated intramuscularly at the left calf with RABV and monitored daily up to 30 dpi. (A) Survival rate of mice (n = 10) inoculated with rToyo and Toyohashi at doses of 102 FFU/head. (B) Survival rate of mice (n = 5) inoculated with rCVS and CVS at doses of 106 FFU/head. Statistical differences in the survival rate between rToyo, Toyohashi and Yokohama groups, and between rCVS and CVS groups were examined by a log-rank test. ns, not significant (p ≧ 0.05). (C) Incubation periods of mice inoculated with rToyo, Toyohashi and Yokohama at doses of 102 FFU/head, and mice inoculated with rCVS and CVS at doses of 106 FFU/head. Each column shows the date of disease onset in each mouse (means ± standard deviation). *Number of sick/infected mice.
Distribution of RABV-infected cells in the brain after peripheral infection
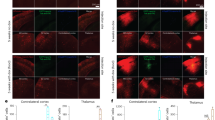
It has been reported that the mice infected with street strains showed a relatively limited distribution of the virus in the brain compared to the CVS strain5. To compare the extent of virus distribution in the brain after peripheral infection, in situ hybridization (ISH) analysis was performed on the brain of mice in the terminal phase after intramuscular inoculation. Viral mRNA positive cells were distributed locally through the brain of the mice infected with rToyo as well as Toyohashi and were identified intensively in the medial cortex, peripheral thalamus and hypothalamus, midbrain, cerebral cortex, medulla and cerebellum, whereas the signals were less in the hippocampus, caudate putamen, ventral striatum, center of the thalamus and olfactory bulb (Fig. 3A,B). Thus, the recombinant and original Toyohashi strains displayed similar distribution in the mouse brain.
Distribution of viral mRNA in the brains of RABV-infected mice. Six-week-old male ddY mice were inoculated intramuscularly with 103 FFU of rToyo or Toyohashi. Mice inoculated with PBS were included as controls. (A) Representative images of the distribution of RABV N mRNA (red) in the whole brains at the terminal stage of infection (6 dpi). The scale bars correspond to 1000 μm. (B) Magnified fields of the cerebral neocortex and hippocampus, cerebellum of Toyohashi (left panel) and rToyo (right panel)-infected mice. The scale bars correspond to 200 μm.
Live-cell imaging of the intracellular trafficing of the viral P protein using fluorescent reporter expressing RABV
Using our established RG system for the Toyohashi strain, we generated a recombinant virus expressing the viral P protein fused with the fluorescent reporter mCherry protein (rToyo-P-mCherry). A plasmid for rToyo-P-mCherry was constructed by inserting the mCherry gene into the 3′-terminal region of P gene in pToyo plasmid (Fig. 4A). Sanger sequencing confirmed that P and mCherry genes were combined into a single coding sequence by inserting mCherry gene at the intended site of the viral P gene (Fig. 4B).
Generation of fluorescent protein expressing rToyo-P-mCherry for live-cell imaging. (A) Schematic representation of the plasmid construction of rToyo-P-mCherry, a recombinant Toyohashi strain carrying the fluorescent reporter mCherry gene at the 3′-terminus of the viral P gene. (B) Sanger sequencing chromatograms of C terminus region of rToyo and rToyo-P-mCherry. (C) Images of fluorescent foci in NA cells at 4 dpi with RABVs. Cells were strained with anti-RABV N antibody (green) and Hoechst 33342 (blue). The scale bars on the images indicate 100 μm. (D) Intracellular trafficking of Toyohashi P protein. SK-N-SH cells were infected with rToyo-P-mCherry and then monitored by live-cell time-lapse imaging from 14 to 26 hpi. Fluorescence signals from P-mCherry were visualized in the red color. Yellow arrowheads indicate fusion between inclusion bodies. Scale bars indicate 20 μm and the time post-infection is displayed in the lower left corner of each panel. All the images have been extracted from Supplementary Movie and are shown at 10-min intervals.
Expression of mCherry protein from cells infected with rToyo-P-mCherry was assessed by fluorescence focus assay in NA cells. The fluorescent foci positive for mCherry were also positive for immunofluorescent staining with anti-RABV N antibody (Fig. 4C). We compared the growth kinetics between rToyo-P-mCherry and wild-type Toyohashi strains and found that the virus titer of rToyo-P-mCherry was slightly lower than that of wild-type viruses (Supplementary Fig. S2).
We next attempted to monitor the intracellular movement of viral P protein in SK-N-SH cells inoculated with rToyo-P-mCherry by live-cell time-lapse imaging analysis. At 14 h post infection (hpi), Negri body-like structures were observed as several spherical structures positive for mCherry (Fig. 4D). Subsequently, the structures increased the number, and fused with each other to form large spheres as time progressed (Supplementary Movie 1), consistent with a previous study using live-cell imaging analysis of CVS-infected cells23,24. Finally, the cells also accumulated Negri body-like structures of various sizes in the cytosol.
Discussion
In this study, we aimed to establish an RG system for analysis of the street Toyohashi strain. Infectious recombinant virus was successfully recovered from cloned cDNA. To the best of our knowledge, this is the first study to establish an RG system for RABV street strain belonging to SEA4 subclade circulating only in the Philippines. Previous studies have reported that serial passage of RABV street strains adapted the viruses to proliferate efficiently in cultured cells and decreased viral neuroinvasiveness in mice, exhibiting properties similar to those of the fixed strains25,26. In order to recover the recombinant RABV from cDNA clones, the virus was required to establish multi-round infection and proliferate in cultured cells, which may select variant virus with nucleotide mutation(s) in the genome and different properties from the original strain. However, we demonstrated that the growth ability of rToyo in cultured cells and the pathogenicity of rToyo in mice were almost identical to those of the original Toyohashi strain (LC812291) (Fig. 1). These results indicated that our RG system successfully recovers the Toyohashi strain without impairing the virological properties of the street strain.
RABV street strains are characterized by high neuroinvasiveness and a long and inconsistent incubation period5. In this study, we investigated the phenotypic properties of the Toyohashi strain. Mouse myoblast G-8 cells permitted the infection and replication of the Toyohashi strain, consistent with a previous study which showed that another RABV street strain isolated from field-infected foxes can proliferate in myotubes which were differentiated from myoblasts (Fig. 1B)27. Based on the LD50 values, we estimate that the neuroinvasiveness of the Toyohashi strain is comparable to that of another street strain, Yokohama, and apparently higher than that of the fixed strain CVS (Table 1). The median incubation period of the Toyohashi strain in mice was similar to that of the Yokohama strain and more than 10 days longer than that of the CVS strain (Fig. 2C). Taken together, these results indicate that the Toyohashi strain is a typical street strain causing a long and variable incubation period with high neuroinvasiveness.
It has been reported that street strains spread in a limited fashion within the brain, whereas fixed strains infection is observed throughout the brain5. Supporting this observation, our ISH analysis also identified viral RNA-positive cells in specific regions and not throughout the entire brain mice following i.m. infection with the Toyohashi strain (Fig. 3). Notably, the localization of infected cells in the brain of Toyohashi-infected mice was similar to that of the brain of mice infected with RABV street strain 1088, which showed prominent infection in the cerebral neocortex and less infection in the hippocampus5. All of the findings described above indicate that the Toyohashi strain has a lower growth ability in the central nervous system than the CVS strain.
Negri body-like structures are a cytoplasmic inclusion body consisting of viral N, P, L proteins and viral RNA and act as a platform for viral gene transcription and genome replication23,28. The physical properties of Negri body-like structures have been analyzed by live-cell imaging and fluorescence recovery after photobleaching analysis with recombinant RABV fixed strains expressing fluorescent protein-tagged P protein24,28. Following these studies, we generated rToyo-P-mCherry, a recombinant Toyohashi strain expressing P protein C-terminally fused to the mCherry fluorescent protein. Our live-cell time-lapse imaging showed that mCherry-tagged P proteins forms dynamic and globular structures in the cytosol of cells infected with rToyo-P-mCherry (Fig. 4D, Supplementary Movie 1). To our knowledge, this is the first study to observe the kinetics of Negri body-like structures of a RABV street strain by live-cell imaging. Future comparative analysis of the dynamics of Negri body-like structures of street and fixed strains may provide new insights into RABV replication.
In conclusion, we have established a RG system for the Toyohashi strain and successfully obtained a recombinant virus without impairment of the properties of the street strain. Generation of rToyo-P-mCherry expressing viral protein tagged with a fluorescent protein offers the tool for tracking viral protein and the development of Negri body-like structures in cells. This RG system will be a powerful tool for manipulating the genome of the Toyohashi strain to investigate the molecular basis of infection and pathogenesis of the RABV street strain which is currently prevalent in the Philippines.
Materials and methods
Ethics statement
All animal experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee of Hokkaido University (Approval number 19-0014 and 24-0002). All methods were carried out in accordance with relevant guidelines and regulations for animal experiments in Hokkaido University (National University Corporation Hokkaido University Regulations on Animal Experimentation). This study is reported in accordance with ARRIVE guidelines.
Cells
Baby hamster kidney cells stably expressing T7 RNA polymerase (BHK/T7-9) cells were established previously29. Mouse neuroblastoma (NA) cells were kindly gifted by Dr. McMorris, The Wistar Institute, USA. BHK/T7-9, NA and human neuroblastoma (SK-N-SH) cells (RCB0426, RIKEN BioResource Center, Japan) were cultured in Eagle’s minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS). SK-N-SH cells were maintained in type I collagen-coated plates. Mouse muscle myoblast (G-8) cells (CRL-1456; ATCC, Virginia, USA) were cultured in high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 10% horse serum. All cells were cultivated at 37 °C in the presence of 5% CO2.
Viruses
The master stock of RABV Toyohashi P2I-2M (GenBank LC812291) was obtained from the original RABV Toyohashi strain (GenBank LC619707)9 by passaging twice in the brains of suckling mice and twice on NA cells. RABV Yokohama and rCVS strains were as described previously30,31. The working stocks of RABVs were propagated in NA cells or suckling mouse brains in all experiments.
Plasmid construction
To obtain a plasmid vector pToyo carrying the full-length cDNA of Toyohashi, a total of three cDNA fragments covering the entire viral genome of Toyohashi (11,922 nt) were amplified by reverse transcription (RT)-polymerase chain reaction (PCR) from viral genomic RNA and cloned into a pUC19 plasmid vector containing the T7 promoter and HDV ribozyme cDNA16,17. To obtain a plasmid vector of pToyo-P-mCherry, the mCherry gene fragment was amplified from pmCherry Vector by PCR and inserted at the 3′-terminus of P gene in pToyo plasmid. The resulting plasmids pToyo and pToyo-P-mCherry were transformed into Escherichia coli HST08 cells (Takara Bio, Japan).
Recovery and propagation of rRABVs
Recombinant RABVs were recovered from viral cDNA encoding plasmids as previously described29,30,32. Briefly, BHK/T7-9 cells were co-transfected with pToyo or pToyo-P-mCherry, and helper plasmids pT7IRES-RN, -RP and -RL using TransIT-LT1 (Mirus, Madison, USA). The recovered recombinant RABV (rRABV) clones in the culture supernatants were collected at 5 days post-transfection and propagated twice in NA cells. To confirm the insertion of mCherry gene in virus genome of rToyo-P-mCherry, a cDNA fragment (nucleotide positions from 2258 to 3049 in the genome of Toyohashi) was amplified by RT-PCR and directly sequenced by conventional Sanger sequencing methods.
Virus titration by focus forming assays and immunofluorescence staining
Monolayers of NA cells in 24 well-plates were inoculated with tenfold serially diluted specimens. Following adsorption for 1 h at 37 °C, cells were cultured in overlay medium (MEM containing 2% FBS and 0.5% methylcellulose). After 4 days post-infection, cells were fixed with 4% paraformaldehyde. Cells were stained with fluorescein isothiocyanate (FITC)-labeled anti-RABV N antibody (Fujirebio, Japan) and Hoechst 33342 (Invitrogen, Massachusetts, USA). Infected cells were observed under a fluorescence microscope (IX73, Olympus, Japan) and virus titers were calculated as focus-forming units per milliliter (FFU/mL). The fluorescent images were obtained by CellSens Imaging Software (Olympus).
Growth kinetics
NA and G-8 cells in 24 well-plates were inoculated with Toyohashi and rToyo at a multiplicity of infection (MOI) of 0.01. Cell culture supernatants were collected at 24, 48, 72 and 96 hpi and subjected to titration by the focus forming assay as described above.
Animal experiments
Six-week-old, male ddY mice (Japan SLC Inc., Japan) were inoculated intramuscularly into the left calf muscle with different doses of RABVs in 0.1 ml of PBS. For survival analysis, the body weight was monitored daily up to 30 dpi. The humane endpoint was established as > 20% body weight loss or a moribund state due to severe paralysis. The LD50 of each virus strain was calculated based on methods previously described33. The median survival time was calculated using GraphPad Prism (version 10.1.0, GraphPad Software, San Diego, USA). For ISH, mice were inoculated intramuscularly with 103 FFU of Toyohashi or rToyo. The brains were collected from mice showing > 10% body weight loss at 7–15 dpi and fixed in 10% buffered formalin.
In situ hybridization (ISH)
Fixed brains were embedded in paraffin and sectioned at 4 μm. ISH was performed using the RNAscope 2.5HD Detection Reagent-RED (Advanced Cell Diagnostics, California, USA) according to the manufacturer's instructions. Briefly, brain tissue sections were baked in a dry oven at 60 °C for 1 h and deparaffinized. To block endogenous peroxidases, tissue sections were pretreated with RNAscope H2O2 solution for 10 min at room temperature. The tissue sections were treated with RNAscope Target Retrieval Reagent for 15 min at 98–102 °C and RNAscope Protease Plus Reagent for 20 min at 40 °C. The RNAscope Probe-V-RABV-N-C1 targeting the Toyohashi N RNA region was designed and manufactured by Advanced Cell Diagnostics (Cat. No. 1305721-C1). Tissues were counterstained with hematoxylin, and images were captured using a Nikon ECLIPSE 80i light microscope (Nikon, Japan).
Live-cell time-lapse imaging
SK-N-SH cells in glass base dishes were infected with rToyo-P-mCherry in MEM supplemented with 10% FBS. The cells were placed in a stage top incubation chamber at 37 °C and 5% CO2 (Tokai Hit, Japan). Live-cell time-lapse imaging was carried out from 14 to 26 hpi using a BZ-X800 fluorescence microscope (KEYENCE, Japan).
Statistical analysis
All statistical analysis was performed using GraphPad Prism (version 10.1.0). A Welch’s t-test was employed for the comparison of two groups at multiple time points. A Log-rank (Mantel-cox) test was used for the survival analysis.
Data availability
The nucleotide sequence of RABV Toyohashi P2I-2M is available in the GenBank repository (accession number: LC812291). All other data generated during this study are included in this published article.
References
WHO Expert Consultation on rabies. World Health Organization. WHO expert consultation on rabies: third report. World Health Organization. https://iris.who.int/handle/10665/272364 (2018).
Gan, H. et al. Global burden of rabies in 204 countries and territories, from 1990 to 2019: Results from the Global Burden of Disease Study 2019. Int. J. Infect. Dis. 126, 136–144 (2023).
Bonaparte, S. C., Moodie, J., Undurraga, E. A. & Wallace, R. M. Evaluation of country infrastructure as an indirect measure of dog-mediated human rabies deaths. Front. Vet. Sci. 10, 1147543 (2023).
Lépine, P. On the evolution of fixed strains of rabies virus. J. Hyg. (Lond) 38, 180–184 (1938).
Takahashi, T. et al. Genetic and phenotypic characterization of a rabies virus strain isolated from a dog in Tokyo, Japan in the 1940s. Viruses 12, 914 (2020).
Faber, M. et al. A single amino acid change in rabies virus glycoprotein increases virus spread and enhances virus pathogenicity. J. Virol. 79, 14141–14148 (2005).
Yamaoka, S. et al. Involvement of the rabies virus phosphoprotein gene in neuroinvasiveness. J. Virol. 87, 12327–12338 (2013).
Okada, K. et al. Roles of the rabies virus phosphoprotein isoforms in pathogenesis. J. Virol. 90, 8226–8237 (2016).
Nosaki, Y. et al. Fourth imported rabies case since the eradication of rabies in Japan in 1957. J. Travel Med. 28, taab151 (2021).
Yuson, M., Manalo, D. L., Miranda, M. E. G., Hampson, K. & Telmo, S. Rabies in the greater manila area and region IV-B of the Philippines and the potential impact of age-targeted dog vaccination. One Health Dog-mediat. Rabies Elimin. Asia A Collect. Local Exp. 99–114 (2023) https://doi.org/10.1079/9781800622975.0009.
Faber, M. et al. Identification of viral genomic elements responsible for rabies virus neuroinvasiveness. Proc. Natl. Acad. Sci. U. S. A. 101, 16328–16332 (2004).
Ben Khalifa, Y. et al. The matrix protein of rabies virus binds to RelAp43 to modulate NF-κB-dependent gene expression related to innate immunity. Sci. Rep. 6, 39420 (2016).
Virojanapirom, P., Yamada, K., Khawplod, P., Nishizono, A. & Hemachudha, T. Increased pathogenicity of rabies virus due to modification of a non-coding region. Arch. Virol. 161, 3255–3261 (2016).
Tian, Q. et al. Phosphoprotein gene contributes to the enhanced apoptosis induced by wild-type rabies virus GD-SH-01 in vitro. Front. Microbiol. 8, 1697 (2017).
Nolden, T. et al. Reverse genetics in high throughput: Rapid generation of complete negative strand RNA virus cDNA clones and recombinant viruses thereof. Sci. Rep. 6, 23887 (2016).
Isomura, M., Yamada, K., Noguchi, K. & Nishizono, A. Near-infrared fluorescent protein iRFP720 is optimal for in vivo fluorescence imaging of rabies virus infection. J. Gen. Virol. 98, 2689–2698 (2017).
Takahashi, T. et al. Establishment of a reverse genetics system for rabies virus strain Komatsugawa. J. Vet. Med. Sci. 84, 1508–1513 (2022).
Virojanapirom, P. et al. Molecular analysis of the mutational effects of Thai street rabies virus with increased virulence in mice after passages in the BHK cell line. Arch. Virol. 157, 2201–2205 (2012).
Delmas, O. et al. Genomic diversity and evolution of the lyssaviruses. PLoS One 3, e2057 (2008).
Luo, Y. et al. Complete genome sequence of a highly virulent rabies virus isolated from a rabid pig in South China. J. Virol. 86, 12454–12455 (2012).
Troupin, C. et al. Large-scale phylogenomic analysis reveals the complex evolutionary history of rabies virus in multiple carnivore hosts. PLoS Pathog. 12, e1006041 (2016).
Zhang, L. et al. Large-scale phylogenetic analysis reveals genetic diversity and geographic distribution of rabies virus in South-East and South Asia. Infect. Genet. Evol. 113, 105472 (2023).
Nikolic, J. et al. Negri bodies are viral factories with properties of liquid organelles. Nat. Commun. 8, 58 (2017).
Nikolic, J., Civas, A., Lama, Z., Lagaudrière-Gesbert, C. & Blondel, D. Rabies virus infection induces the formation of stress granules closely connected to the viral factories. PLoS Pathog. 12, e1005942 (2016).
Yamada, K. et al. Serial passage of a street rabies virus in mouse neuroblastoma cells resulted in attenuation: Potential role of the additional N-glycosylation of a viral glycoprotein in the reduced pathogenicity of street rabies virus. Virus Res. 165, 34–45 (2012).
Nitschel, S. et al. Point mutations in the glycoprotein ectodomain of field rabies viruses mediate cell culture adaptation through improved virus release in a host cell dependent and independent manner. Viruses 13, 1989 (2021).
Tsiang, H. & Tsiang, H. Rabies virus infection of myotubes and neurons as elements of the neuromuscular junction. Rev. Infect. Dis. 10, S733–S738 (1988).
Lahaye, X. et al. Functional Characterization of Negri Bodies (NBs) in rabies virus-infected cells: Evidence that NBs are sites of viral transcription and replication. J. Virol. 83, 7948–7958 (2009).
Ito, N., Takayama, M., Yamada, K., Sugiyama, M. & Minamoto, N. Rescue of rabies virus from cloned cdna and identification of the pathogenicity-related gene: Glycoprotein gene is associated with virulence for adult mice. J. Virol. 75, 9121–9128 (2001).
Anindita, P. D. et al. Generation of recombinant rabies viruses encoding NanoLuc luciferase for antiviral activity assays. Virus Res. 215, 121–128 (2016).
Tamashiro, H., Matibag, G. C., Ditangco, R. A., Kanda, K. & Ohbayashi, Y. Revisiting rabies in Japan: Is there cause for alarm?. Travel Med. Infect. Dis. 5, 263–275 (2007).
Itakura, Y. et al. Glu333 in rabies virus glycoprotein is involved in virus attenuation through astrocyte infection and interferon responses. iScience 25, 104122 (2022).
Harshbarger, K. The American. Antioch. Rev. 72, 546–558 (2014).
Acknowledgements
This study was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI under Grant numbers 23H02376 and 23KJ0053; the Japan Agency for Medical Research and Development (AMED) Grant number JP243fa627005; the Japan Science and Technology Agency (JST) Moonshot R&D Grant number JPMJMS2025; the Ministry of Health, Labour and Welfare Grant number JPMH19HA1008; the World-leading Innovative and Smart Education (WISE) Program from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan under Grant number 1801.
Author information
Authors and Affiliations
Contributions
N.K., M.S. designed the study. N.K., Y.I., K.I., T.A., Y.O., M.S. performed the experiments. N.K., Y.I., M.S. analysed the data. H.S., Y.O., M.S. supervised this study. M.H., S.I., K.M., N.I. contributed to critical resources and methodology. N.K. W.W.H., H.S., M.S. wrote the original draft. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Video 1.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kawaguchi, N., Itakura, Y., Intaruck, K. et al. Reverse genetic approaches allowing the characterization of the rabies virus street strain belonging to the SEA4 subclade. Sci Rep 14, 18509 (2024). https://doi.org/10.1038/s41598-024-69613-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-69613-y
Keywords
This article is cited by
-
Development of viral infectious clones and their applications based on yeast and bacterial artificial chromosome platforms
Molecular Biomedicine (2025)
-
Exploring the role of infected keratinocytes during rabies virus infection
npj Viruses (2025)