Abstract
Temporomandibular joint disorders are prevalent causes of orofacial discomfort. Diagnosis predominantly relies on assessing the configuration and positions of temporomandibular joint components in magnetic resonance images. The complex anatomy of the temporomandibular joint, coupled with the variability in magnetic resonance image quality, often hinders an accurate diagnosis. To surmount this challenge, we developed deep learning models tailored to the automatic segmentation of temporomandibular joint components, including the temporal bone, disc, and condyle. These models underwent rigorous training and validation utilizing a dataset of 3693 magnetic resonance images from 542 patients. Upon evaluation, our ensemble model, which combines five individual models, yielded average Dice similarity coefficients of 0.867, 0.733, 0.904, and 0.952 for the temporal bone, disc, condyle, and background class during internal testing. In the external validation, the average Dice similarity coefficients values for the temporal bone, disc, condyle, and background were 0.720, 0.604, 0.800, and 0.869, respectively. When applied in a clinical setting, these artificial intelligence-augmented tools enhanced the diagnostic accuracy of physicians, especially when discerning between temporomandibular joint anterior disc displacement and osteoarthritis. In essence, automated temporomandibular joint segmentation by our deep learning approach, stands as a promising aid in refining temporomandibular joint disorders diagnosis and treatment strategies.
Similar content being viewed by others
Introduction
Temporomandibular joint disorders (TMDs) constitute a diverse range of pathologies affecting the temporomandibular joint (TMJ) and its affiliated anatomical structures1,2,3. These conditions present with various clinical manifestations, encompassing cephalic pain, facial asymmetry, tinnitus, and other related symptoms4,5,6,7. TMDs have garnered attention healthcare industry because of their prevalence, ranking as the second most frequently observed musculoskeletal affliction. Between 5 and 12% of the U.S. population is affected by one or more of these disorders, resulting in an annual economic burden of approximately $4 billion4,8. Furthermore, a significant 20–25% of the U.S. demographic reports TMJ-associated symptoms, underscoring the critical public health implications, with an alarming rate of almost one million new cases emerging annually9,10,11.
Magnetic resonance imaging (MRI) is a fundamental noninvasive diagnostic tool for evaluating various TMDs, including internal disc derangements and osteoarthritic changes12,13,14,15,16,17,18. For accurate diagnosis, the precise delineation of the TMJ components on MR images is essential, with TMJ segmentation focusing on the spatial attributes and morphology of the components, which include the temporal bone, disc, and condyle19. However, manual delineation presents challenges because of ambiguities in disc visualization and the poor contrast in magnetic resonance (MR) images19. Clinicians should meticulously examine not only disc displacements, joint effusions, chronic inflammation, and abnormalities in the mandibular condyle, but also the microarchitectural integrity of soft tissues, including the disc, temporal bone, and mandibular condyle, during the diagnostic process20,21. They rely heavily on their expertise in this process. However, the aforementioned limitations of MRI, combined with the dynamic nature of TMJ MRI, can make it difficult to determine the boundaries of each anatomical component, particularly the disc, condyle, and temporal bone. Additionally, the morphology of TMJ components can change based on disease progression, further complicating the segmentation process and increasing the risk of errors22,23.
Recently, the dentistry field has seen a rise in artificial intelligence adoption15,24,25,26. However, TMJ component segmentation in MR images remains challenging due to issues with contrast and visibility. Few studies have addressed TMJ segmentation comprehensively. Some excluded the disc23, others relied on manual methods27, and many automated approaches focused only on the disc, neglecting the condyle and temporal bone28,29. Additionally, several studies used limited regions of interest (ROI) or binary segmentation techniques, which do not meet the diverse diagnostic needs of TMDs28.
This study investigated a method for the automated segmentation of the three primary TMJ components, namely the temporal bone, disc, and condyle, and validated its efficacy using ensemble deep learning methodologies specifically tailored to multi-class segmentation (Fig. 1). Additionally, our method incorporates not only convolutional neural networks, which use convolution operations commonly employed in previous TMJ segmentation studies, but also Vision Transformer models, which excel at capturing global context and enhancing feature representation. By combining these models in an ensemble learning framework, our approach significantly improves the accuracy and robustness of TMJ segmentation. This innovative method provides clinicians with a powerful diagnostic tool that facilitates rapid and precise TMD diagnoses, streamlining clinical decision-making processes and optimizing therapeutic interventions across a spectrum of TMD presentations (Fig. 2).
Deep learning pipeline for magnetic resonance (MR) image preprocessing and segmentation. The figure presents a two-part workflow for medical image analysis. On the left, the 'Image preprocessing' segment outlines the initial treatment of a MR image. It begins with manual annotation on a region of interest (ROI), followed by three key steps: cropping to focus on the ROI, applying Contrast Limited Adaptive Histogram Equalization (CLAHE) for contrast enhancement, and performing data augmentation to enhance the robustness and variability of the image data. The right-hand section, labeled 'Deep learning pipeline,' displays the processed MRI image being input into a suite of deep learning models for further analysis. These models include DeepLabV3Plus, OCRNet with ResNet101, UPerNet with ResNet101, UPerNet combined with the Swin Transformer, and SegFormer. Each contributes to the analysis, and their outputs are integrated using an ensemble method. This integration leads to the creation of a 'Prediction mask,' which visualizes the segmentation outcome, providing a detailed delineation of the ROI against the background image.
Results
This study delineated the outcomes of segmenting the TMJ using six salient deep learning architectures, including five baseline models and one ensemble model. During training, with epochs capped at 50, a customary behavior was observed across the five baseline models: the validation loss initially decreased and then increased after reaching the epoch with the minimum loss. The epoch corresponding to this lowest validation loss varied depending on the model. To prevent overfitting and ensure optimal performance, we selected the model weights at the point of minimum validation loss for each model. This approach allowed us to use the most effective model state, avoiding the effects of overfitting observed in later epochs (Table 1).
In the internal validation, the DSC values for the temporal bone, disc, condyle, and background varied, with ranges of 0.847–0.868, 0.694–0.733, 0.882–0.910, and 0.947–0.952, respectively (Table 2a).
In the external validation, the DSC values for the temporal bone, disc, condyle, and background fluctuated within ranges of 0.692–0.733, 0.561–0.607, 0.771–0.800, and 0.858–0.870, respectively (Table 2b).
Both the temporal bone and mandibular condyle consistently demonstrated high DSC value, emphasizing the reliability of the model's performance for these components. By contrast, the disc component consistently had lower DSC values. This trend of disparities among the components persisted even in the external validation, underlining the consistent nature of the model's behavior across different datasets.
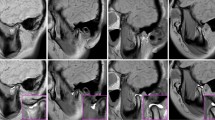
Figure 3 visually illustrates the outcomes of segmenting the TMJ components. The demarcated regions for the temporal bone and condyle were predominantly accurate, with minimal aberrations observed peripherally. The segmentation of the disc component appeared to be slightly indistinct; however, its spatial placement was accurately discerned, demonstrating that this model could play a significant role in diagnosing disc displacement. Concurrently, the background delineation was notably accurate. Additionally, Fig. 4 depicts a representative TMJ disc displacement, indicating the relative positioning of the joint's components, while Fig. 5 exhibits a TMJ osteoarthritis example, underlining the condylar morphology but with a slightly subdued edge definition in comparison to the reference.
Utilizing MRI slices alone, Doctor A diagnosed TMJ anterior disc displacement (ADD) and osteoarthritis with 53.33% accuracy. After introducing the AI-generated masks, the accuracy increased to 73.33% for TMJ ADD and 60% for osteoarthritis. Similarly, Doctor B's accuracy improved from 50 to 80% for TMJ ADD and from 50 to 66.67% for osteoarthritis. Doctor C's accuracy increased from 76.67 to 90% for TMJ ADD, with a minor improvement for osteoarthritis from 60 to 66.67%. When using the AI-predicted masks with the MRI, the precision levels for the diagnosis revisions were as follows: Doctor A at 85.71%, Doctor B at 93.75%, and Doctor C at 77.78%.
Discussion
Our study introduced a novel approach to the segmentation of TMJ components in MR images using multi-class deep learning, which showed significant advancements over previous methodologies. Our model demonstrated strong performance with DSC values of 0.855, 0.711, 0.894, and 0.949 for the respective components. This improved performance can be attributed to the innovative use of ensemble learning and Vision Transformers. The ensemble learning approach combines the strengths of multiple models, enhancing robustness and accuracy. Specifically, it leverages the complementary capabilities of convolutional neural networks (CNNs) and Vision Transformers (ViTs). CNNs are highly effective in capturing local features due to their convolutional operations, which is crucial for detailed structure segmentation in MR images. On the other hand, ViTs, with their attention mechanisms, excel at capturing global context and enhancing feature representation, which is particularly beneficial for accurately distinguishing between closely located anatomical structures within the TMJ. By integrating these models, the ensemble learning approach ensures a more comprehensive analysis of the image data, leading to higher segmentation accuracy. The combination of local detail capture by CNNs and global context understanding by ViTs allows our model to effectively handle the inherent challenges of TMJ component segmentation, such as contrast variability and visibility issues. In summary, the success of our model in achieving high DSC values for TMJ component segmentation is largely due to the synergistic effects of ensemble learning and the advanced capabilities of Vision Transformers in enhancing feature representation and model performance.
Our research achieved significant advancements in the domain of MR imaging analysis, especially in the context of the TMJ. The study contributions can be categorized into two primary domains. First, the inherent challenges of MR imaging often arise from the weak signal intensity of certain anatomical structures, notably the TMJ disc. To address this issue, we implemented a contrast enhancement algorithm. This approach not only accentuated the visibility of the TMJ disc but also sharpened the demarcation of the associated TMJ structures, including the temporal bone and condyle. An integral aspect of this methodology was the meticulous calibration of the hyperparameters that governed the algorithm's performance. The optimal values for these hyperparameters were identified using a comprehensive grid search technique. Note that any imprecision in this calibration process could amplify noise levels within the MR image, underlining the importance of rigorous parameter tuning. Second, a well-acknowledged challenge in medical imaging is the requirement for substantial datasets to facilitate efficient deep learning. To address the challenges associated with amassing extensive medical data, our research leveraged transfer learning. By drawing upon insights from pretrained models on expansive datasets, we achieved commendable efficiency, even with the relatively limited data pertaining to TMJ MR imaging. Moreover, we incorporated data augmentation techniques to enhance our model's resilience to data limitations. By integrating transformations including random rotations and shifts into our original dataset, we fortified the model's robustness and generalizability. Notably, our empirical observations revealed that the amalgamation of transfer learning and data augmentation led to a performance increase of 5–10% compared to models without these enhancements.
The results underscore the potential of AI-integrated diagnostic approaches in the realm of TMDs. Upon analyzing physician comparative outcomes, a consistent improvement in diagnostic rates was evident upon the inclusion of the AI-predicted mask, which suggests that the AI-assisted approach was reliable in refining clinical decision-making processes.
For a condition such as TMJ ADD, the increase in the diagnostic rate when using the AI-predicted mask was noteworthy. All the participating physicians demonstrated increased diagnostic accuracies, indicating the potential of the AI model to assist in diverse clinical scenarios. A similar positive trajectory was evident in osteoarthritis diagnosis, emphasizing the broad applicability of our segmentation models.
When considering corrections made after mask integration, the high precision rates indicated that the AI's impact was not confined to increasing the number of diagnoses but also included improvements in accuracy. This is critical in a clinical setting where diagnostic precision directly influences patient care and outcomes.
From an anatomical perspective, the introduced segmentation models offer a refined method for assessing TMJ components through MRI. By facilitating automated and meticulous insights, clinicians can gain a deeper understanding of the intricate relationships between the TMJ components. Consequently, the proposed model would be an invaluable supplementary tool, enhancing the diagnostic process for TMDs and ensuring a higher standard of patient care.
Our research had certain limitations. The discrepancy between the DSC measures for the internal and external validations could be seen as a manifestation of overfitting. When a model is overfit to internal data, it may capture noise or very specific patterns that do not generalize well to external datasets. To overcome this issue, the model's generalization capabilities must be enhanced. Obtaining diverse data from a variety of patients, ensuring a broad spectrum of cases and scenarios, would be effective. By training the model on such augmented and varied datasets, it could learn more general features rather than over-relying on specific patterns from a limited dataset. Consequently, the model’s performance will improve in the external validation, making it more robust and applicable across different patient cohorts.
Prior research on TMJ segmentation relied on a bounding box method to determine the ROI by marking corner coordinates that encompassed the entire region. In practical clinical scenarios, an automated system is needed that can seamlessly extract an ROI that captures all the vital TMJ components from a comprehensive MR image. Therefore, a promising direction for future studies would be the development of a TMJ ROI detection model and its integration with our TMJ segmentation model, to establish an end-to-end automated system for clinical application.
This study presented an innovative AI-driven methodology for the autonomous segmentation of the three integral components of the TMJ. Employing this systematic segmentation, clinicians could considerably streamline the diagnostic process for various TMJ disorders. Furthermore, our AI-anchored TMJ segmentation model delivers consistent and precise predictions, which, when combined with conventional clinical assessments, could bolster the fidelity of the eventual diagnoses. In essence, the convergence of AI and digital dentistry offers promising avenues for enhanced diagnostic precision and efficacy in addressing dental and TMJ disorders.
Materials and methods
Study participants
The institutional review boards of Ewha Womans University Mokdong Hospital (EUMC 2020-03-012-003) and Yonsei University Gangnam Severance Hospital (3-2020-0025) approved this study, and we affirm that all experiments were performed in accordance with relevant guidelines and regulations. Ewha Womans University Mokdong Hospital Institutional Review Board and Yonsei University Gangnam Severance Hospital Institutional Review Board absolved the need for informed consent given the negligible risks posed to the study participants. Rigorous measures were instituted to safeguard the confidentiality of the patient information throughout the investigation. Patients exhibiting MRI artifacts and those with divergent annotations from disparate clinicians were methodically excluded from the study.
Two distinct datasets were employed for the research's analytical objectives: Dataset-E and Dataset-Y (Table 1). Dataset-E was judiciously utilized for the development and internal testing of the deep learning algorithms, while Dataset-Y was used for the external validation. Dataset-E encapsulated data from 479 participants (age spectrum: 15–68 years; average age ± standard deviation: 32 ± 13 years; male constituency: 41%) accrued from Ewha Womans University Mokdong Hospital (Hospital-E), South Korea, spanning the period from April 2005 to April 2020. Dataset-Y encompassed data from 63 participants (age spectrum: 19–77 years; average age ± standard deviation: 33 ± 13 years; male constituency: 40%) gathered from Yonsei University Gangnam Severance Hospital (Hospital-Y), South Korea, within the identical time frame.
Magnetic resonance imaging (MRI) protocol
For Dataset-E, MRI slices were performed with a Siemens Avanto 1.5 Tesla Magnetom apparatus furnished with dual 3-inch surface coils. The sequences recorded and used for training, validation, and testing were T1-weighted images. The specified acquisition parameters included a repetition time (TR) of 400.00 ms, an echo time (TE) of 15.00 ms, a field of view (FOV) spanning 160 × 160 mm2, an acquisition matrix of 256 × 256 pixels, and a slice thickness of 3 mm. Subsequent to the acquisition, the MR slices were digitized to resolutions of either 2048 × 2048 or 1024 × 1024 pixels and archived using the Standard JPEG 1992 format.
For Dataset-Y, MR images were procured using a 3.0-T Magnetom scanner (Achieva; Philips Medical Systems, Best, The Netherlands) complemented by 3-inch surface coils. The sequences recorded and used for training, validation, and testing were T1-weighted images. The delineated acquisition parameters included a TR of 450.00 ms, a TE of 15.00 ms, an FOV measuring 120 × 120 mm2, an acquisition matrix of 240 × 240 pixels, and a slice thickness of 3 mm. After their acquisition, the MR slices were digitized to a resolution of 512 × 512 pixels and archived using the Standard JPEG 1992 format.
Each participant's MRI volumetric data comprised a total of 40 slices, divided into 20 with an open-mouth configuration and 20 with a closed-mouth configuration. A meticulous criterion was devised to ascertain the quintessential loci within the MRI volume to optimally visualize the TMJ component. Consequently, a subset of eight MR slices pertinent to these loci was extracted. This extraction comprised two MR slices each for the right joint in the closed-mouth posture, left joint in the closed-mouth posture, right joint in the open-mouth posture, and left joint in the open-mouth posture. Slices manifesting suboptimal visibility of the TMJ component or those in which segmentation masks were infeasible because of image clarity concerns were categorically excluded. Moreover, MR slices exhibiting conspicuous artifacts were judiciously excised to maintain the integrity and quality of the dataset.
Datasets
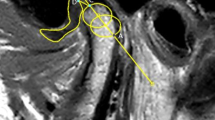
The comprehensive datasets consisted of paired entities: an MR image juxtaposed with its corresponding segmentation mask, serving as the ground truth. For Dataset-E, the TMJ components were meticulously delineated within a circumscribed ROI by three clinicians (Group 1: 16, 8, and 7 years of experience) in oral and maxillofacial surgery and were reviewed by a single expert (JW Kim). For Dataset-Y, six clinicians (Group 2: 14, 13, 10, 9,7, and 6 years of experience) in oral and maxillofacial surgery participated in the delineation, and a single expert (JY Kim) reviewed the dataset. The ROI was characterized as the minimal spatial expanse adequately encompassing the temporal bone, disc, and condyle, which are represented in red, green, and yellow in the figures, respectively.
Dataset-E was partitioned into a development set and an intramural test set. The development set was meticulously split into training subset for model training and validation subset used for the periodic assessment of model performance, culminating in the selection of the optimal model. To ensure a balanced distribution, patients from Dataset-E were randomly assigned at the patient-level to the training subset, validation subset, and intramural test set at a 7:1:2 ratio. For specificity, the development set of Dataset-E included 383 patients, equivalent to 702 joints, for a total of 2,660 images. The intramural test set comprised 96 patients, which translated to 160 joints and 532 images. By contrast, Dataset-Y was distinctively configured as an extramural test set, with data from 63 patients, for a total of 126 joints and 501 images.
Model development and test
Figure 2 represents the whole process. Beginning with an MR image, a specific "region of interest (ROI)" is manually annotated. This ROI undergoes sequential preprocessing, including cropping to isolate the region, contrast enhancement using CLAHE, and data augmentation. The processed image is then fed into five deep learning models, including DeepLabV3plus paired with resnet10130, OCRNet31, UPerNet paired with resnet10132, UPerNet integrated with the Swin transformer33, and SegFormer34. The outputs of these models are combined using an ensemble method, culminating in a final "prediction mask" that showcases the segmentation results.
Image preprocessing
We utilized a computer-assisted approach to extract the ROI from sagittal MR images with an automated cropping algorithm, which enhanced the efficiency.
Owing to the low contrast of MR images, the contrast-limited adaptive histogram equalization (CLAHE) technique35 was implemented for the training subset utilizing the Albumentations Python library36. The optimized hyperparameters for CLAHE included a clip limit of 4 and grid size of 8.
We incorporated image augmentation to diversify the training dataset and prevent overfitting. We used random rotations within a range of \(-{15}^{^\circ }\text{to}+{15}^{^\circ }\) and shifts of up to 10% for both the x and y dimensions. This augmentation was repeated five times.
Lastly, all the ROI MR images and corresponding masks were standardized to \(640\times 640\) pixels. These methods collectively supported a robust deep learning model for medical imaging.
Development and test
This research was dedicated to devising and scrutinizing deep learning models tailored to the precise segmentation of the TMJ components within MRI slices. Convolutional neural networks (CNNs) have been the cornerstone for image analysis. However, transformers, which are equipped with self-attention mechanisms, have challenged this norm because of their performance in specific contexts.
Five architectures were rigorously evaluated: three grounded in CNN—DeepLabV3plus paired with resnet101, OCRNet, and UPerNet paired with resnet101. Simultaneously, two leveraged the nuances of transformers: UPerNet integrated with the Swin transformer, and SegFormer. The UPerNet with Swin transformer combines the hierarchical feature extraction capabilities of UPerNet with the powerful global attention mechanism of the Swin transformer, allowing for improved spatial understanding and segmentation accuracy. SegFormer, on the other hand, utilizes a transformer encoder-decoder architecture to provide efficient and effective segmentation by capturing both local and global features, ensuring precise delineation of TMJ components. The segmentation challenge was framed as a pixel classification task, where each pixel was either earmarked for a specific TMJ component or categorized as background.
During training, models underwent iterative evaluation across epochs. At each epoch's culmination, the model's efficacy on the training subset was gauged against the validation subset, which guided the fine-tuning of the hyperparameters. These were configured at a learning rate of 0.01, momentum of 0.9, weight decay of 0.005, batch size of 4, and span of 50 epochs. PyTorch37 facilitated model development and was powered by four NVIDIA TESLA V100 GPUs.
The cross-entropy loss function was employed to quantify the alignment between the model predictions and ground truth. This function, which is traditionally preferred for multi-category classification, was used to determine the discrepancies. Stochastic gradient descent was selected to train the model by minimizing this loss.
By recognizing the constraints of our dataset size, transfer learning was harnessed. This entailed the initialization of our models with weights from architectures pretrained on the PASCAL VOC 2012 dataset, followed by fine-tuning on our TMJ dataset. This strategy, which is especially potent when grappling with limited data, ensured a robust foundation for our models.
Lastly, ensemble techniques were integrated to assess the compounded strength of our five trained models. Ensemble methodologies strengthen model robustness and accuracy by harmonizing predictions across different architectures38. In our approach, we utilized a majority voting system for individual model outputs. This method ensures that the ensemble's prediction is based on the consensus of the majority of models, enhancing accuracy and reducing the likelihood of errors from any single model. By combining the strengths of both CNN-based models and transformer-based models, the ensemble method capitalizes on their complementary capabilities. The majority voting system ensures that the final prediction benefits from the detailed local feature extraction of CNNs and the global context understanding of transformers, resulting in more robust and precise TMJ component segmentation.
Evaluation and statistical analysis
To assess the effectiveness of our models in predicting image segmentation masks, we employed a test dataset and evaluated the similarity between the predicted and ground truth masks using the Dice similarity coefficient (DSC), sensitivity, and specificity. The DSC was calculated using the following equation:
where TP represents the number of true positive pixels, FP represents the number of false positive pixels, and FN represents the number of false negative pixels. The DSC values ranged from 0 to 1, where a higher value indicated a better segmentation performance.
Sensitivity, also known as the true positive rate, measures the proportion of actual positives that are correctly identified by the model. We calculated sensitivity using the following equation:
where TP represents the number of true positive pixels, and FN represents the number of false negative pixels. Sensitivity values range from 0 to 1, with higher values indicating better performance in correctly identifying pixels belonging to the object of interest.
Specificity, also known as the true negative rate, measures the proportion of actual negatives that are correctly identified by the model. We calculated specificity using the following equation:
where TN represents the number of true negative pixels, and FP represents the number of false positive pixels. Specificity values range from 0 to 1, with higher values indicating better performance in correctly identifying pixels not belonging to the object of interest.
To evaluate the statistical robustness of the model's outcomes, a nonparametric bootstrap methodology was employed to derive the 95% confidence intervals. This entailed a stochastic selection of N instances from the test dataset through a replacement mechanism, iterated 1000 times. Subsequently, from the accrued distribution of these 1000 bootstrap samples, the 95% confidence intervals for the DSC, sensitivity, and specificity performance indicators were ascertained by discerning the range demarcated by the 2.5th and 97.5th percentiles. This methodological framework provided a comprehensive estimation of the inherent variability associated with the model's performance.
Physician comparative analysis: diagnoses with and without AI-predicted masks
TMJ diagnostic procedure
For evaluating the clinical utility of AI-predicted masks in both the TMJ ADD and TMJ osteoarthritis diagnoses, we involved a separate group of 3 external experts (Group 3), who had not participated in the annotation process of either dataset. Group 3 consisted of three experienced clinicians from the department of oral and maxillofacial surgery, with 21, 19, and 17 years of experience respectively. They assessed a set of 30 joints randomly selected from Dataset-Y, the external test dataset. This was done to ensure a rigorous and unbiased evaluation of the AI-predicted masks.
The MRIs for the initial diagnostic round were the same MRIs used in the following diagnostic round with the AI masks. This was to maintain consistency and ensure that any observed improvement in diagnostic performance was due to the integration of AI-predicted masks.
Calculation procedure
The increase in the diagnostic rate was calculated by determining the difference between the numbers of diagnoses made with and without the AI-predicted mask and then dividing it by the total number of joints.
To gauge the precision of the correct diagnoses made using the AI-predicted masks, we determined the percentage of cases where the diagnosis was altered after mask inclusion and turned out to be correct.
Ethical approval
This study was approved by the institutional review boards of two hospitals, Ewha Womans University Mokdong Hospital (EUMC 2020-03-012-003) and Yonsei University Gangnam Severance Hospital (3-2020-0025).
Data availability
This study was approved by the Institutional Review Board of Ewha Womans University Mokdong Hospital and Yonsei University Gangnam Severance Hospital. The data generated and analyzed during this study are protected because of patient privacy concerns. Some data may be available from the corresponding author upon reasonable request, subject to certain restrictions.
References
Bi, R. et al. Divergent chondro/osteogenic transduction laws of fibrocartilage stem cell drive temporomandibular joint osteoarthritis in growing mice. (2023).
Li, J., Ma, K., Yi, D., Oh, C.-D. & Chen, D. Nociceptive behavioural assessments in mouse models of temporomandibular joint disorders. Int. J. Oral Sci. 12, 26 (2020).
Lai, Y. et al. Kindlin-2 loss in condylar chondrocytes causes spontaneous osteoarthritic lesions in the temporomandibular joint in mice. Int. J. Oral Sci. 14, 33 (2022).
Jacobs, T. & Sawaengkit, P. National institute of dental and craniofacial research efficacy trials of bionator class II treatment: A review. Angle Orthod. 72, 571–575 (2002).
Tanaka, E., Detamore, M. & Mercuri, L. Degenerative disorders of the temporomandibular joint: etiology, diagnosis, and treatment. J. Dent. Res. 87, 296–307 (2008).
Kerstens, H. C., Tuinzing, D. B. & van der Kwast, W. A. Temporomandibular joint symptoms in orthognathic surgery. J. Cranio-Maxillofac. Surg. 17, 215–218 (1989).
Kuchler-Bopp, S. et al. Temporomandibular joint damage in K/BxN arthritic mice. Int. J. Oral Sci. 12, 5 (2020).
Schiffman, E. et al. Diagnostic criteria for temporomandibular disorders (DC/TMD) for clinical and research applications: Recommendations of the international RDC/TMD consortium Network and orofacial pain special interest group. J. Oral Facial Pain Headache 28, 6 (2014).
Ingawale, S. & Goswami, T. Temporomandibular joint: disorders, treatments, and biomechanics. Ann. Biomed. Eng. 37, 976–996 (2009).
Isong, U., Gansky, S. A. & Plesh, O. Temporomandibular joint and muscle disorder-type pain in US adults: The National Health Interview Survey. J. Orofac. Pain 22, 317 (2008).
Detamore, M. S. & Athanasiou, K. A. Structure and function of the temporomandibular joint disc: Implications for tissue engineering. J. Oral Maxillofac. Surg. 61, 494–506 (2003).
Rao, V. M. et al. Altered condylar morphology associated with disc displacement in TMJ dysfunction: observations by MRI. Magn. Reson. Imaging 8, 231–235 (1990).
Roh, H.-S., Kim, W., Kim, Y.-K. & Lee, J.-Y. Relationships between disk displacement, joint effusion, and degenerative changes of the TMJ in TMD patients based on MRI findings. J. Cranio-Maxillofac. Surg. 40, 283–286 (2012).
Oezmen, Y., Mischkowski, R. A., Lenzen, J. & Fischbach, R. MRI examination of the TMJ and functional results after conservative and surgical treatment of mandibular condyle fractures. Int. J. oral Maxillofac. Surg. 27, 33–37 (1998).
Helenius, L. et al. Clinical, radiographic and MRI findings of the temporomandibular joint in patients with different rheumatic diseases. Int. J. Oral Maxillofac. Surg. 35, 983–989 (2006).
Larheim, T. A. Role of magnetic resonance imaging in the clinical diagnosis of the temporomandibular joint. Cells Tissues Organs 180, 6–21 (2005).
Rafaat, M. & Shalan, A. Role of magnetic resonance imaging in assessment of temporomandibular joint internal derangement. Benha Med. J. 37, 369–381 (2020).
Kim, J.-Y., Kim, D., Jeon, K. J., Kim, H. & Huh, J.-K. Using deep learning to predict temporomandibular joint disc perforation based on magnetic resonance imaging. Sci. Rep. 11, 6680 (2021).
Smirg, O., Liberda, O., Smekal, Z. & Sprlakova-Pukova, A. MRI slice segmentation and 3D modelling of temporomandibular joint measured by microscopic coil. Measurement Sci. Rev. 12, 74 (2012).
Cui, S.-J. et al. Chronic inflammation deteriorates structure and function of collagen fibril in rat temporomandibular joint disc. Int. J. Oral Sci. 11, 2 (2019).
Shrivastava, M., Battaglino, R. & Ye, L. A comprehensive review on biomarkers associated with painful temporomandibular disorders. Int. J. Oral Sci. 13, 23 (2021).
Alkhader, M. et al. Diagnostic performance of magnetic resonance imaging for detecting osseous abnormalities of the temporomandibular joint and its correlation with cone beam computed tomography. Dentomaxillofacial Radiol. 39, 270–276 (2010).
Liu, Y., Lu, Y., Fan, Y. & Mao, L. Tracking-based deep learning method for temporomandibular joint segmentation. Ann. Transl. Med. 9, 467 (2021).
Jader, G. et al. in 2018 31st SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI). 400–407 (IEEE).
Hao, J. et al. Toward clinically applicable 3-dimensional tooth segmentation via deep learning. J. Dental Res. 101, 304–311 (2022).
Wang, H. et al. Multiclass CBCT image segmentation for orthodontics with deep learning. J Dental Res. 100, 943–949 (2021).
Al-Saleh, M. A. et al. Three-dimensional assessment of temporomandibular joint using MRI-CBCT image registration. PLoS One 12, e0169555 (2017).
Nozawa, M. et al. Automatic segmentation of the temporomandibular joint disc on magnetic resonance images using a deep learning technique. Dentomaxillofacial Radiol 51, 20210185 (2022).
Ito, S. et al. Automated segmentation of articular disc of the temporomandibular joint on magnetic resonance images using deep learning. Sci. Rep. 12, 221 (2022).
Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F. & Adam, H. in Proceedings of the European Conference on Computer Vision (ECCV). 801–818.
Yuan, Y., Chen, X. & Wang, J. in Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, August 23–28, 2020, Proceedings, Part VI 16. 173–190 (Springer).
Xiao, T., Liu, Y., Zhou, B., Jiang, Y. & Sun, J. in Proceedings of the European Conference on Computer Vision (ECCV). 418–434.
Liu, Z. et al. in Proceedings of the IEEE/CVF International Conference on Computer Vision. 10012–10022.
Xie, E. et al. SegFormer: Simple and efficient design for semantic segmentation with transformers. Adv. Neural Inform. Process. Syst. 34, 12077–12090 (2021).
Pizer, S. M. et al. Adaptive histogram equalization and its variations. Comput Vis. Gr. Image Process. 39, 355–368 (1987).
Buslaev, A. et al. Albumentations: Fast and flexible image augmentations. Information 11, 125 (2020).
Paszke, A. et al. Pytorch: An imperative style, high-performance deep learning library. Adv. Neural Inform. Process. Syst. 32 (2019).
Sagi, O. & Rokach, L. Ensemble learning: A survey. Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery 8, e1249 (2018).
Acknowledgements
This work was supported by an Institute of Information & Communications Technology Planning & Evaluation (IITP) grant funded by the Korea government (MSIT) (No. RS-2022-00155966, Artificial Intelligence Convergence Innovation Human Resources Development (Ewha Womans University)) The work of J. Choi and J.W. Kim was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (Grant Number: HI23C0162).
Author information
Authors and Affiliations
Contributions
K. Yoon and J. Choi contributed to the conception, design, data analysis, and interpretation, as well as the drafting and critical revision of the manuscript; J.W. Kim contributed to the conception, design, data acquisition, and interpretation, as well as the drafting and critical revision of the manuscript; J.Y. Kim, S.J. Kim, and J.K. Huh contributed to the data acquisition and interpretation, as well as the drafting and critical revision of the manuscript. All the authors gave their final approval and agreed to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yoon, K., Kim, JY., Kim, SJ. et al. Multi-class segmentation of temporomandibular joint using ensemble deep learning. Sci Rep 14, 18990 (2024). https://doi.org/10.1038/s41598-024-69814-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-69814-5
Keywords
This article is cited by
-
Diagnostic accuracy of artificial intelligence models for temporomandibular joint anomalies on MRI: a systematic review and meta-analysis
BioMedical Engineering OnLine (2026)
-
3D CSFA-UNet: a unified attention-driven deep learning framework for accurate knee MRI segmentation and osteoarthritis severity classification
Scientific Reports (2026)
-
An attention-based transfer learning model for diagnosing subluxation in temporomandibular joint panoramic radiographs
Scientific Reports (2025)