Abstract
The current detection method for Chikungunya Virus (CHIKV) involves an invasive and costly molecular biology procedure as the gold standard diagnostic method. Consequently, the search for a non-invasive, more cost-effective, reagent-free, and sustainable method for the detection of CHIKV infection is imperative for public health. The portable Fourier-transform infrared coupled with Attenuated Total Reflection (ATR-FTIR) platform was applied to discriminate systemic diseases using saliva, however, the salivary diagnostic application in viral diseases is less explored. The study aimed to identify unique vibrational modes of salivary infrared profiles to detect CHIKV infection using chemometrics and artificial intelligence algorithms. Thus, we intradermally challenged interferon-gamma gene knockout C57/BL6 mice with CHIKV (20 µl, 1 X 105 PFU/ml, n = 6) or vehicle (20 µl, n = 7). Saliva and serum samples were collected on day 3 (due to the peak of viremia). CHIKV infection was confirmed by Real-time PCR in the serum of CHIKV-infected mice. The best pattern classification showed a sensitivity of 83%, specificity of 86%, and accuracy of 85% using support vector machine (SVM) algorithms. Our results suggest that the salivary ATR-FTIR platform can discriminate CHIKV infection with the potential to be applied as a non-invasive, sustainable, and cost-effective detection tool for this emerging disease.
Similar content being viewed by others
Introduction
Chikungunya virus (CHIKV) is an arbovirus transmitted to humans mainly by Aedes aegypti and Aedes albopictus mosquitoes. CHIKV was first isolated in 1952 in Tanzania and this alphavirus caused intermittent outbreaks in several countries in all continents1, except Antarctica2. The CHIKV has a 12 kb positive-sense RNA genome that encodes two polyproteins. It is subdivided into four non-structural proteins (nsP1, nsP2, nsP3, nsP4) and other five structural proteins (C, E3, E2, 6 K, and E1)3. The E2 glycoprotein is a viral surface protein important to cell binding into host cells, while the interaction of E2 viral protein with several receptors in host cells involved in this viral entry include clathrin, epidermal growth factor receptor substrate 15 (Eps15), heparan sulfate, keratan sulfate, chondroitin sulfate, and dermatan sulfate for CHIKV entry4.
CHIKV causes an inflammatory musculoskeletal disease in humans with symptoms, such as arthralgia, arthritis, and myalgia3. The CHIKV infection leads also to a febrile disease known as chikungunya fever (CHIKF), and their common symptom is polyarthralgia mainly in the large joints (knees and elbows) and peripheral joints (ankles, wrists, and phalanges)2,5. The acute stage of CHIKV occurs one week after infection and the chronic stage can occur for months to years in humans. Acute fever and polyarthralgia are the main factors involved in the symptomatology of CHIKV infection1,6. Data from the European Centre for Disease Prevention and Control (ECDC) indicates 383,357 reported cases of CHIKV infection worldwide, mainly reported in Brazil, India, Paraguay, Guatemala, and Thailand7.
The specific tests for CHIKV diagnostics are mainly based on blood samples and the type of test used depends on the time of collection after symptoms oncet8. These tests are based on viral RNA detection9 using Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)10,11,12, or based on serological tests with detection of specific IgM and IgG antibodies against CHIKV using enzyme-linked immunosorbent assays (ELISA) and/or rapid immunochromatographic assys13. The salivary detection of viral components has become an important alternative after the COVID-19 pandemic14,15,16,17. Salivary detection can be considered an interesting choice in the diagnosis of systemic diseases. Salivary composition includes organic and inorganic elements, such as electrolytes, peptides, proteins, carbohydrates, nucleic acids, and pathogens including viruses. Saliva permits auto-collection using a relatively fast, painless, effortless, and safe non-invasive collection method; saliva is also more convenient to store compared to blood18. Some studies confirmed the presence of CHIKV in saliva, where RNA was detected in 58.3% of saliva samples during the 1st-week post symptoms onset19. Besides, saliva samples collected from several acute CHIKV patients with hemorrhagic manifestations also contain viral RNA and infectious virus20.
The application of Fourier-transform infrared coupled with Attenuated Total Reflection (ATR-FTIR) spectroscopy can contribute to the detection of several emerging viral infections. ATR-FTIR is a powerful analytical tool based on mid-infrared region21 for quantitative and qualitative analysis of biofluids22,23,24,25,26,27,28, and it is an outstanding spectroscopic technique21. ATR-FTIR can provide information concerning the characterization of proteins, lipids, nucleic acids, and carbohydrates26. The region of the electromagnetic spectrum comprises the 4000–400 cm−1 range. In biological samples, the range between 1800 and 900 cm−1 is known as the biofingerprint region29,30.
Here, we tested the hypothesis that unique spectral salivary biomarkers can be differentially expressed in the saliva of CHIKV-infected mice to be used as salivary infrared markers for CHIKV detection. In this context, the present study aimed to identify a salivary infrared profile to detect CHIKV signatures and to evaluate the accuracy of chemometrics analysis and machine learning algorithms based on infrared spectra obtained in a sustainable and portable biophotonic platform.
Materials and methods
Animals
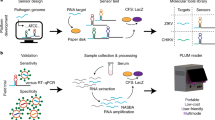
Interferon-gamma gene knockout C57/BL6 male mice (29.5 g and ~ 2 months-old) were divided into: vehicle (n = 7) and CHIKV (n = 6). The animals were maintained under standard conditions (22 ± 2 °C, 55% ± 5% humidity, and 12-h light/dark cycles, light on at 5 AM) and were allowed free access to standard diet and water at the Center for Bioterism and Experimentation (REBIR) at the Federal University of Uberlandia (UFU). Mice were inoculated once in the left rear footpad with (20 µL, 1 × 105 PFU/mL) and control mice received vehicle [physiological saline, sodium chloride 0.9%] in a sterile solution (20 µL) (Fig. 1). All experimental procedures were approved by the Ethics Committee for Animal Research at UFU (License CEUA-UFU #023/2021), according to Ethical Principles adopted by the Brazilian College of Animal Experimentation (COBEA) and ARRIVE guidelines.
Serum, and saliva sample collection
At Day 3, due to the peak of viremia31, both vehicle and CHIKV animals were anesthetized with 12 mg/kg of xylazine and 80 mg/kg of ketamine, and subsequently, these mice received pilocarpine (2 mg/kg, i.p.) to stimulate salivary secretion (Fig. 1). The saliva was collected for 5 min, and the samples were stored at -80 °C until further analysis. Besides, the blood was also collected, and serum was separated by centrifuging at 2500 rpm for 15 min in a refrigerated centrifuge Thermo Scientific Heraeus Megafuge 16R. The animals were euthanized by exsanguination, a process involving the insertion of a needle into the heart cavity, followed by blood withdrawal.
RNA extraction, cDNA synthesis, and qPCR amplification
RNA extraction of serum samples was performed automated on the Maxwell 16 RSC Extraction System (Promega, USA) using the Maxwell RSC Viral Total Nucleic Acid Purification kit (Promega, USA)32. After this process, viral RNA was used for cDNA transcription using GoScript Reverse Transcriptase (Promega, USA) with OligoDT (Promega, USA). The samples were amplified in the Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, USA) using the GoTaq qPCR master mix A6002 (Promega, USA) following the manufacturer’s instructions, and employing the primers design to target the CHIKV E region: forward (5′-GTCACATACCACCCTCG-3′) and reverse (5′-TGYCTCTTAGGGGACACATATACCT-3′)33.
ATR-FTIR spectroscopy, spectra data procedures, and statical analysis
Spectra of saliva samples were recorded using the portable Agilent Cary 630 FTIR Spectrometer connected to micro-attenuated total reflectance (ATR) mode in the range of 4000 to 650 cm−1. Spectra were acquired with 2 cm−1 spectral resolution and 64 scans (co-adds) per spectrum for each sample. The polypropylene microtube with saliva was inserted in a vortex for 3 min to homogenate the sample, and 2 μl of saliva was inserted directly dried at ATR-crystal. After 5 min using airflow directly to the samples, each dried salivary spectrum was individually collected34.
The salivary infrared profile was processed using Orange 3.3.5 software. For the generation of the mean spectra, the baseline was corrected by the Rubberband method followed by Min–Max normalization to avoid errors throughout sample preparation and spectra analysis. Then, the infrared profile truncated compost for the lipidic region (3050–2800 cm−1) and the fingerprint region (1800–900 cm−1) to input data for chemometrics analysis as principal component analysis (PCA)27. In the PCA analyses, each vibrational mode is projected onto a new subspace identified exclusively by the main principal components to obtain reduced-dimensional data even though preserving data variation partially. In this exploratory (unsupervised) analysis of spectra data, the principal component was assembled with scores (deviation in sample orientation) and loadings (variance in the wavenumber direction)35.
Predictive analysis method
The infrared spectral data analysis was divided into two stages: pre-processing and classification. Pre-processing consisted of aggregation, attribute selection, and data transformation. The spectral readings of each sample were performed in aggregation, the spectral data was truncated using the lipidic region (3050–2800 cm−1) associated with the biofingerprint region (1800–900 cm−1). Then, the Savitzky-Golay smoothing filter was applied followed by a first-order derivative and amide I normalization to each spectrum35.
The classification was tested with state-of-the-art machine learning algorithms and linear discriminant analysis of feature extraction coupled with discriminant analysis tools. The Linear Discriminant Analysis (LDA) and Support Vector Machine (SVM) algorithms were selected based on better results during model training. LDA classifies sets of patients or measurements into groups by projecting the original data onto a lower dimensional space. SVM is a supervised learning technique that fits a hyperplane through data projected in a high-dimensional space, allowing for the classification of spectra or patient data. A five-times stratified cross-validation was applied to analyze the predictive performance of the LDA and SVM algorithms. In this procedure, the samples are divided into 5 disjoint sets and, in each run, 4 sets are used as training data, and 1 set is exclusively used as the test, resulting in a total of 5 runs. To measure the results obtained, three predictive performance metrics were used: sensitivity, specificity, and accuracy. The sensitivity or true positive rate is the proportion of positive samples (animals infected with CHIKV) that were correctly classified, and the specificity or true negative rate is the proportion of negatives (control animals or vehicles) that were correctly classified. The accuracy is defined as the percentage of total samples correctly classified considering true and false negatives27,30.
Shapley Additive Explanations (SHAP) is an effective model to explain artificial intelligence algorithms. Accordingly, SHAP calculates the contribution of each vibrational mode to the target value. The SHAP value of each vibrational within the best-performing artificial intelligence model can help to identify which features contribute more to discrimination between positive samples (animals infected with CHIKV) and negative samples (control animals or vehicles)36.
Results
The body weight of CHIKV-infected mice (CHIKV: 29 ± 0.9 g) was similar (p > 0.05) to vehicle mice. CHIKV RNA replication was present in the serum of CHIKV mice. According to the qPCR protocol, the molecular testing showed a positive result for CHIKV detection in the serum samples (CT: 29.63 ± 0.4). The other vehicle samples were negative for CHIKV molecular detection (CT > 37).
Salivary infrared profile
The mean infrared original spectrum of saliva in the fingerprint region (1800–800 cm−1) collected from the vehicle and CHIKV mice indicates a superposition of salivary components as proteins, lipids, DNA/RNA, and carbohydrates (Fig. 2).
Subsequently, the PCA was used to reduce the dimensional space by comparing the infrared spectral changes in vehicle and CHIKV mice with truncated spectra including the lipidic region (3050–2800 cm−1) associated with the fingerprint region (1800-900 cm−1). These spectra displayed significant differences in the salivary spectra of vehicle mice and CHIKV mice. Two of three principal components (PCs) named PC1 and PC2 explained 71.2% of cumulative variance. Both PC1 (42.7% explained variance) and PC2 (28.5% explained variance) are represented in the scores plot (72.8% explained variance in Fig. 3A). The PCA loadings to PC1, PC2, and PC3 are represented in Fig. 2B explained 86.9%. We highlighted the higher absolute coefficients − 0.045, − 0.0625, and − 0.075 represented by these subsequent vibrational modes at 1583 cm−1, 1355 cm−1, and 1035 cm−1 in PC1, respectively. For PC2, the coefficients 0.0765, − 0.041, and − 0.076 represent the vibrational modes 1553 cm−1, 1494 cm−1 and 1015 cm−1 respectively; finally, the vibrational modes for PC3, 1651 cm−1, 1619 cm−1, 1525 cm−1 and 1350 cm−1 has the coefficients − 0.071, 0.064, − 0,098 and 0,058, respectively.
Discrimination analysis
To classify and perform predictive analysis of salivary spectral samples from the vehicle and CHIKV mice, artificial intelligence tools were applied, most notably machine learning with LDA and SVM (Table 1). The results obtained indicate that the best discriminatory analysis when comparing vehicle mice to CHIKV mice was using SVM algorithms. The best pre-processing applied was obtained by the Savitzky-Golay. The most favorable outcome was using the spectral range of 3050–2800 cm−1 in combination with 1800–900 cm−1. These pre-processed infrared spectra showed a sensitivity of 83%, specificity of 86%, and accuracy of 85% between vehicle and CHIKV mice.
Finally, we applied Shapley Additive Explanations (SHAP), a state-of-the-art approach to explain the output of machine learning models. The technique works by associating optimal credit allocation with local explanations using Shapley values from game theory and other extensions. Here we adopted SHAP to obtain a better understanding about the vibrational modes that contributed more to the better predictive results achieved by SVM. The score value of each feature based on the SVM algorithm under pre-processed spectra obtained by Savitzky-Golay indicates the main vibrational modes responsible for the discrimination of vehicle and CHIKV mice. As an outcome, the main vibrational modes to discriminate vehicle and CHIKV samples were 1636 cm−1, 1635 cm−1, 1660 cm−1, 1661 cm−1, 1637 cm−1, 1662 cm−1, 1659 cm−1, 2966 cm−1, and 1638 cm−1 (Fig. 4). The molecular assignment suggestions of each vibrational mode indicated by the SHAP feature analysis—SVM were described in Table 2.
Discussion
The application of portable, non-invasive, and reagent-free platforms for CHIKV detection using saliva samples could be a robust alternative to improve the detection of CHIKV, which is crucial to the adequate management of CHIKV-infected patients. In this context, to the best of our knowledge, the current study is pioneer for the translational applicability of this sustainable biophotonic platform to the salivary detection of CHIKV infection in a murine animal model.
Currently, the diagnosis of CHIKV infection depends on clinical, epidemiological, and laboratory criteria. The signs and symptoms of CHIKV infection should be distinguished from other suspect arbovirus, especially, alphaviruses diseases with similar clinical outcomes40. The detection of viral RNA can be performed using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) or a one-step PCR assay, which is an expensive tool with a rapid indication of viral load in clinical samples or cell culture supernatant41. The detection of immune response to CHIKV is based on serological methods such as enzyme-linked assays (ELISA), indirect immunofluorescence assays (IFA), hemagglutination inhibition (HI), and micro-neutralization (MNt). These immune response assays do not detect viral components and the detection 5 days after symptoms onset depends on the CHIKV strain40,42. Considering the high costs and the long time to deliver results related to the diagnostics of CHIKV infection with current tests, it is imperative to evaluate novel reagent-free, non-invasive, and rapid platforms to detect CHIKV infection in the acute phase using saliva samples.
Supporting our hypothesis some infrared vibrational modes of saliva were capable to discriminate CHIKV-infected mice than control mice. PCA analysis was applied to evaluate the salivary spectra from CHIKV-infected and vehicle mice. The PCA explained 86.9% of the cumulative variance in three Principal Components (PCs). For each component, the vibrational modes 1583 cm−1, 1355 cm−1 and 1035 cm−1 in PC1; 1553 cm−1, 1494 cm−1 and 1015 cm−1 in PC2; and 1651 cm−1, 1619 cm−1, 1525 cm−1, 1350 cm−1 in PC3 were fundamental for an explanation. These vibrational modes are related to changes in salivary components including the β-sheet structure of amide I in proteins and Stretching C-H of lipids. CHIKV is an enveloped alphavirus with a positive-sense RNA genome and around 12 kb in length43. Although ATR-FTIR spectrometry can detect accurately nucleic acids profile44,45,46, this was not selected with SHAP models for CHIKV infection detection. The detection of proteins and lipids in the saliva of animals with CHIKV is likely linked to the host’s immune response, which can promote an increase in the presence of immunoglobulins and an altered expression of lipids due to CHIKV infection.
The application of machine learning algorithms in the infrared signature indicates SVM as best performed algorithm coupled in a pre-processing Savitzky-Golay achieved an accuracy of 85%, with a sensitivity of 83%. This data indicates the potential of this biophotonic platform to be used in the CHIKV infection, with similar sensitivity with antigen detection in rapid tests, where the sensitivity can vary depending on antigen levels47. Recently, the application of machine learning algorithms in infrared biofluid spectra was used to discriminate viral diseases such as COVID-19, Dengue, and Zika Virus18,48,49, representing a great potential for diagnostic perspectives on viral infection.
Here, we also described the ten best-performing spectral biomarkers based on the SHAP values for CHIKV infection detection. These vibrational modes are related to salivary proteins and salivary lipids, suggesting changes in the expression of these organic compounds in the saliva of CHIKV-infected mice. The β-sheet structure of Amide I indicates potential changes in salivary peptides and/or salivary proteins related to CHIKV infection. This molecular structure is related to a powerful infrared signal with a high sensitivity to detect changes in molecular geometry50 and is assembled by the stretching vibrations of the C = O (70%—85%) and C-N groups (10%—20%)51. Taking into consideration, the similarity of salivary secretion and CHIKV-infection mechanisms in both animal models and humans, it suggests this salivary vibrational mode is a potential candidate for CHIKV detection in humans.
To the best of our knowledge, this study is pioneer in the development of CHIKV detection using a sustainable, reagent-free, and highly reproducible platform using a non-invasive sample. With further development, the development of this approach may provide a novel alternative to detect CHIIKV infection based on a unique analysis of spectral salivary components using artificial intelligence algorithms. This pan-omic spectroscopic salivary analysis associated with an orthogonal approach conducted against a different immunogenic target could provide a novel alternative for this viral detection52.
It is imperative to highlight that ATR-FTIR has been used for biofluids analysis, allowing a rapid results release and open new perspectives for disease detection26,27,34,35. The salivary photonic-based diagnostics proposed here should be tested in large sample cohorts in patients and may unveil the prospect for point-of-care assays by portable infrared spectroscopic devices. Additionally, it is important to highlight that CHIKV diagnosis for RT-qPCR requires high-cost equipment and expensive reagents even on a large scale. In this context, the employment of a label-free ATR-FTIR has robust potential to reduce costs in CHIKV infection detection and reduce the impact of medical waste53.
Despite the promising results of the study, several limitations are recognized, including the small sample size of infected mice and the need to validate the findings in humans to confirm their clinical applicability. However, the number of animals is adequate for the 3Rs principles and ethical guidelines for rodents. The study focused on a single post-infection time point, so longitudinal analyses are required to monitor dynamic variations in salivary biomarkers. Even so, this study indicates the beginning or the basis for carrying out other studies with the use of the technique, where future research should include other viral infections, integrate advanced machine learning algorithms, and explore underlying molecular mechanisms, which could lead to the creation of a universal diagnostic tool. Furthermore, it is crucial to emphasize that the experimental animals in this study were provided with a standardized diet, had ad libitum access to clean water, and were maintained under consistent temperature and humidity conditions throughout the experiment. Consequently, the observed differences can be attributed directly to exposure to CHIKV infection. Collaboration with public health experts would facilitate the practical application of these findings, improving outbreak response strategies.
This proof-of-concept study indicates that salivary infrared signatures are enable to detect CHIKV using a sustainable biophotonic platform. The optimal pattern classification demonstrated a suitable sensitivity of 83%, a specificity of 86%, and an overall accuracy of 85% when employing support vector machine (SVM) algorithms on the infrared signature of saliva samples from CHIKV-infected and vehicle mice. Our findings suggest that the salivary ATR-FTIR platform possesses the capability to effectively discern CHIKV infection, which suggests promising utility as a non-invasive, sustainable, and cost-effective detection tool for addressing the challenges in developing detection strategies for this emerging disease. Additional larger sample sizes in clinical studies should be assessed comparing healthy and CHIKV to indicate the potential of this pan-omic spectroscopic platform using saliva samples as a clinical detection test for CHIKV infection.
Data availability
All data generated or analysed during this study are included in this published article.
References
Ganesan, V. K., Duan, B. & Reid, S. P. Chikungunya virus: Pathophysiology, mechanism, and modeling. Viruses 9, 368 (2017).
Silva, J. V. J. et al. A scoping review of Chikungunya virus infection: Epidemiology, clinical characteristics, viral co-circulation complications, and control. Acta Trop. 188, 213–224 (2018).
Matusali, G. et al. Tropism of the Chikungunya virus. Viruses 11, 175 (2019).
Schnierle, B. S. Cellular attachment and entry factors for Chikungunya virus. Viruses 11, 1078 (2019).
Morrison, T. E. Reemergence of Chikungunya virus. J. Virol. 88, 11644–11647 (2014).
Lum, F.-M. & Ng, L. F. P. Cellular and molecular mechanisms of chikungunya pathogenesis. Antivir. Res. 120, 165–174 (2015).
Chikungunya worldwide overview. European Centre for Disease Prevention and Control https://www.ecdc.europa.eu/en/chikungunya-monthly.
da Cunha, R. V. & Trinta, K. S. Chikungunya virus: Clinical aspects and treatment—A review. Mem. Inst. Oswaldo Cruz 112, 523–531 (2017).
Johnson, B. W., Russell, B. J. & Goodman, C. H. Laboratory diagnosis of Chikungunya virus infections and commercial sources for diagnostic assays. J. Infect. Dis. 214, S471–S474 (2016).
Jalloh, M. A. et al. Seroprevalence of Chikungunya in an asymptomatic adult population in North and South Sulawesi, Indonesia. Am. J. Trop. Med. Hyg. 108, 359–362 (2023).
Xu, Z. et al. Simultaneous detection of Zika, Chikungunya, dengue, yellow fever, West Nile, and Japanese encephalitis viruses by a two-tube multiplex real-time RT-PCR assay. J. Med. Virol. 94, 2528–2536 (2022).
Monteiro, J. D. et al. Epidemiologic and clinical investigations during a Chikungunya outbreak in Rio Grande do Norte State Brazil. PLoS One 15, e0241799 (2020).
Lo Presti, A., Lai, A., Cella, E., Zehender, G. & Ciccozzi, M. Chikungunya virus, epidemiology, clinics and phylogenesis: A review. Asian Pac. J. Trop. Med. 7, 925–932 (2014).
Sabino-Silva, R., Jardim, A. C. G. & Siqueira, W. L. Coronavirus COVID-19 impacts to dentistry and potential salivary diagnosis. Clin. Oral. Investig. 24, 1619–1621 (2020).
Shamsoddin, E. Saliva: A diagnostic option and a transmission route for 2019-nCoV. Evid. Based Dent. 21, 68–70 (2020).
Sri Santosh, T., Parmar, R., Anand, H., Srikanth, K. & Saritha, M. A Review of salivary diagnostics and its potential implication in detection of covid-19. Cureus 12, e7708 (2020).
Kamel, M. et al. Non-invasive detection of SARS-CoV-2 antigen in saliva versus nasopharyngeal swabs using nanobodies conjugated gold nanoparticles. Trop. Med. Infect. Dis. 7, 102 (2022).
Oliveira, S. W. et al. Salivary detection of Zika virus infection using ATR-FTIR spectroscopy coupled with Machine learning algorithms and univariate analysis: A proof-of-concept animal study. Diagnostics (Basel) 13, 1443 (2023).
Musso, D. et al. Detection of Chikungunya virus in saliva and urine. Virol. J. 13, 102 (2016).
Gardner, J. et al. Infectious Chikungunya virus in the saliva of mice Monkeys and humans. PLoS One 10, e0139481 (2015).
Martin, F. L. et al. Distinguishing cell types or populations based on the computational analysis of their infrared spectra. Nat. Protoc. 5, 1748–1760 (2010).
Ojeda, J. J. & Dittrich, M. Fourier transform infrared spectroscopy for molecular analysis of microbial cells. Methods Mol. Biol. 881, 187–211 (2012).
Kazarian, S. G. & Chan, K. L. A. Applications of ATR-FTIR spectroscopic imaging to biomedical samples. Biochim. Biophys. Acta 1758, 858–867 (2006).
de Sousa Marques, A. et al. Feature selection strategies for identification of Staphylococcus aureus recovered in blood cultures using FT-IR spectroscopy successive projections algorithm for variable selection: A case study. J. Microbiol. Methods 98, 26–30 (2014).
Sakudo, A., Suganuma, Y., Sakima, R. & Ikuta, K. Diagnosis of HIV-1 infection by near-infrared spectroscopy: Analysis using molecular clones of various HIV-1 subtypes. Clin. Chim. Acta 413, 467–472 (2012).
Ferreira, I. C. C. et al. Attenuated total reflection-fourier transform infrared (ATR-FTIR) spectroscopy analysis of saliva for breast cancer diagnosis. J. Oncol. 2020, 4343590 (2020).
Caixeta, D. C. et al. Salivary molecular spectroscopy: A sustainable, rapid and non-invasive monitoring tool for diabetes mellitus during insulin treatment. PLoS One 15, e0223461 (2020).
Wood, B. R. et al. Infrared based saliva screening test for COVID-19. Angew. Chem. Int. Ed. 60, 17102–17107 (2021).
Movasaghi, Z. et al. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 43, 134–179 (2008).
Baker, M. J. et al. Using fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 9, 1771–1791 (2014).
Haese, N. N. et al. Animal models of Chikungunya virus infection and disease. J. Infect. Dis. 214, S482–S487 (2016).
Maxwell® RSC 48 Instrument. https://www.promega.com.br/products/lab-automation/automated-dna-rna-extraction-purification-maxwell/maxwell-rsc-48-instrument/?catNum=AS8500.
Simmons, M. et al. Development and validation of a quantitative, One-step, multiplex, real-time reverse transcriptase pcr assay for detection of dengue and Chikungunya viruses. J. Clin. Microbiol. 54, 1766–1773 (2016).
Rodrigues, R. P. et al. Differential molecular signature of human saliva using ATR-FTIR spectroscopy for chronic kidney disease diagnosis. Braz. Dent. J. 30, 437–445 (2019).
Caixeta, D. C. et al. Monitoring glucose levels in urine using FTIR spectroscopy combined with univariate and multivariate statistical methods. Spectrochim. Acta A Mol. Biomol. Spectrosc. 290, 122259 (2022).
Akulich, F., Anahideh, H., Sheyyab, M. & Ambre, D. Explainable predictive modeling for limited spectral data. Chemom. Intell. Lab. Syst. 225, 104572 (2022).
Eckel, R. et al. Characteristic infrared spectroscopic patterns in the protein bands of human breast cancer tissue. Vib. Spectrosc. 27, 165–173 (2001).
Yang, Y. et al. Study of tumor cell invasion by Fourier transform infrared microspectroscopy. Biopolymers 78, 311–317 (2005).
Dovbeshko, G. I., Gridina, N. Y., Kruglova, E. B. & Pashchuk, O. P. FTIR spectroscopy studies of nucleic acid damage. Talanta 53, 233–246 (2000).
Caglioti, C. et al. Chikungunya virus infection: An overview. New Microbiol. 36, 211–227 (2013).
Dash, M., Mohanty, I. & Padhi, S. Laboratory diagnosis of chikungunya virus: do we really need it?. Indian J. Med. Sci. 65, 83–91 (2011).
Cho, B., Kim, J., Cho, J.-E., Jeon, B.-Y. & Park, S. Expression of the capsid protein of Chikungunya virus in a baculovirus for serodiagnosis of Chikungunya disease. J. Virol. Methods 154, 154–159 (2008).
Kendall, C. et al. Structural and phenotypic analysis of Chikungunya virus RNA replication elements. Nucl. Acids Res. 47, 9296–9312 (2019).
Tunçer, S. & Gurbanov, R. Non-growth inhibitory doses of dimethyl sulfoxide alter gene expression and epigenetic pattern of bacteria. Appl. Microbiol. Biotechnol. 107, 299–312 (2023).
Ceylani, T., Teker, H. T., Samgane, G. & Gurbanov, R. Intermittent fasting-induced biomolecular modifications in rat tissues detected by ATR-FTIR spectroscopy and machine learning algorithms. Anal. Biochem. 654, 114825 (2022).
Teker, H. T. et al. Reduced liver damage and fibrosis with combined SCD Probiotics and intermittent fasting in aged rat. J. Cell. Mol. Med. 28, e18014 (2024).
Ujj, B. (CDC/DDPHSS/CSELS/DLS). Diagnostic Sensitivity and Specificity for Clinical Laboratory Testing.
Barauna, V. G. et al. Ultrarapid on-site detection of SARS-CoV-2 infection using simple ATR-FTIR spectroscopy and an analysis algorithm: High sensitivity and specificity. Anal. Chem. 93, 2950–2958 (2021).
Santos, M. C. D., Nascimento, Y. M., Araújo, J. M. G. & Lima, K. M. G. ATR-FTIR spectroscopy coupled with multivariate analysis techniques for the identification of DENV-3 in different concentrations in blood and serum: A new approach. RSC Adv. 7, 25640–25649 (2017).
Khanmohammadi, M. & Garmarudi, A. B. Infrared spectroscopy provides a green analytical chemistry tool for direct diagnosis of cancer. TrAC Trends Anal. Chem. 30, 864–874 (2011).
Kumar, S. et al. Vibrational studies of different human body disorders using FTIR spectroscopy. Open J. Appl. Sci. https://doi.org/10.4236/ojapps.2014.43012 (2014).
Cameron, J. M. et al. A spectroscopic liquid biopsy for the earlier detection of multiple cancer types. Br. J. Cancer 129, 1658–1666 (2023).
Letho, Z. et al. Awareness and practice of medical waste management among healthcare providers in national referral hospital. PLoS One 16, e0243817 (2021).
Acknowledgements
We would like to thank our collaborators at the Rodent Vivarium Network (REBIR-UFU). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Funding
CAPES—Prevention and Combat of Outbreaks, Endemics, Epidemics, and Pandemics (#23038.014934/2020–59), FAPEMIG (#APQ-02872–16, APQ-03613–17, APQ-00476–20, APQ-02148–21, APQ-00410-21), Federal University of Uberlandia and National Institute of Science and Technology in Theranostics and Nanobiotechnology (CNPq Process N.: 465669/2014–0, 403193/2022–2). Sabino-Silva, R received a fellowship from FAU/UFU, CNPq productivity fellowship (306050/2021–8), and PrInt CAPES/UFU.
Author information
Authors and Affiliations
Contributions
M.G.V. and R.S–S. conceived and designed the experiments; M.G.V., G.M.F., R.B.R., R.C.S., M.A.C. and D.C.C. performed the experiments. M.G.V., D.C.C., A.C.M.F., and M.G.C. analyzed the data. A.C.G. and R.S–S. contributed reagents/materials/analysis tools; M.G.V., G.M.F., M.G.C., and R.S–S. wrote the paper; M.G.V and R.S–S. performed the research design.
Corresponding author
Ethics declarations
Competing interests
The authors declare interested in filing a patent with the Brazilian National Institute of Industrial Property (INPI).
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Guevara-Vega, M., Rosa, R.B., Caixeta, D.C. et al. Salivary detection of Chikungunya virus infection using a portable and sustainable biophotonic platform coupled with artificial intelligence algorithms. Sci Rep 14, 21546 (2024). https://doi.org/10.1038/s41598-024-71889-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-71889-z