Abstract
Calcium is involved in vision processes in the retina and implicated in various pathologies, including glaucoma. Rod cells rely on store-operated calcium entry (SOCE) to safeguard against the prolonged lowering of intracellular calcium ion concentrations. Zebrafish that lacked the endoplasmic reticulum Ca2+ sensor Stim2 (stim2 knockout [KO]) exhibited impaired vision and lower light perception-related gene expression. We sought to understand mechanisms that are responsible for vision impairment in stim2 KO zebrafish. The single-cell RNA (scRNA) sequencing of neuronal cells from brains of 5 days postfertilization larvae distinguished 27 cell clusters, 10 of which exhibited distinct gene expression patterns, including amacrine and γ-aminobutyric acid (GABA)ergic retinal interneurons and GABAergic optic tectum cells. Five clusters exhibited significant changes in cell proportions between stim2 KO and controls, including GABAergic diencephalon and optic tectum cells. Transmission electron microscopy of stim2 KO zebrafish revealed decreases in width of the inner plexiform layer, ganglion cells, and their dendrites numbers (a hallmark of glaucoma). GABAergic neuron densities in the inner nuclear layer, including amacrine cells, as well as photoreceptors significantly decreased in stim2 KO zebrafish. Our study suggests a novel role for Stim2 in the regulation of neuronal insulin expression and GABAergic-dependent vision causing glaucoma-like retinal pathology.
Similar content being viewed by others
Introduction
Calcium signaling is implicated in several processes in the retina and underlies various eye pathologies. The dysregulation of calcium homeostasis and calcium signaling is associated with neurodegenerative conditions (reviewed in1,2,3,4). Glaucoma is one such pathology, characterized by damage to the optic nerve, a tract of retinal ganglion cell axons that are covered with myelin5. In glaucoma, retinal ganglion cells (RGCs) undergo numerous alterations, including interruption of the retrograde axonal transport of neurotrophic factors, oxidative and endoplasmic reticulum (ER) stress, and intracellular Ca2+ overload6. This results in a progressive and irreversible decrease in the number of RGCs3,7. The specific mechanism of calcium-dependent glaucoma has not been established. Calpains are calcium-dependent proteases that have been implicated in calcium-dependent glaucoma8,9,10,11,12, in addition to several other causes, such as glutamate receptor overexcitation, high extracellular glutamate levels, changes in the activity of voltage-operated calcium channels, the activation of transient receptor potential 4 (TRPV4), reverse activity of the Na+/Ca2+ exchanger, the activity of protein kinase C (PKc), and mitogen-activated protein kinases (MAPKs)1,4,13,14,15,16. Glutamate excitotoxicity in glaucoma has been linked to an increase in Ca2+ influx into mitochondria17,18,19. In all of these pathways, the pathologies were associated with an increase in Ca2+ levels. However, in rod cells, a low level of Ca2+, which occurs under sustained saturating light conditions, is neurotoxic. To prevent cytosolic levels of Ca2+ from exhaustion, rods use store-operated calcium entry (SOCE)20.
SOCE is the mechanism by which the ER is refilled with Ca2+ and involved in Ca2+ signaling21,22,23. It is activated by ER Ca2+ sensors, called stromal interaction molecules (STIMs), which induce the opening of plasma membrane Ca2+ channels, such as calcium release-activated calcium channel protein Orai and transient receptor potential channels (TRPCs). We reported the presence of STIM and Orai transcripts in cortical neuron cultures of the rat brain24 and subsequently provided evidence of the interaction of endogenous STIM and Orai in neurons and their involvement in SOCE25. The STIM family includes STIM1 and STIM2 proteins, which have different functions and sensitivities to Ca2+ level in the ER26. It is noteworthy that Ca2+ ions that enter cells via SOCE not only replenish intercellular stores but also participate in cellular signaling, for instance, affecting the activity of transcription factors21. High level of STIM2 protein was observed in the central nervous system of rodents and zebrafish, and STIM2 was shown to be crucial for SOCE in mouse neurons. Furthermore, Stim2 KO mice exhibit alterations of the vision system27, including cataracts28, but no changes in retina were reported27. In lenses with cataracts, an increase in intracellular Ca2+ levels was observed, and SOCE was detected in the lens of epithelial cells and retinal pigment epithelial (RPE) cells29,30,31. Although STIM2 has been linked to neurodegeneration32, it remains unknown whether it can be also relevant in the pathogenesis of glaucoma.
Zebrafish (Danio rerio) possess all necessary genes for SOCE, including duplicated stim1 (stim1a and stim1b) and duplicated stim2 (stim2a and stim2b)33. The knockout of either stim2a34 or stim2b35 resulted in increased locomotor activity and anxiety-related thigmotaxis in zebrafish larvae. These larvae exhibited moderate abnormalities in phototactic response, yet their visual-motor response remained normal, indicating that their vision was not affected. However, experiments with stim2a;stim2b-/- double KO (hereinafter referred to as stim2 KO) revealed that Stim2 deficiency in zebrafish resulted in vision impairment, as evidenced by the loss of phototactic behavior, abnormal visual-motor response, alterations of the stratum pigmentosum, as well as thinning of the inner plexiform layer (IPL) and RGCs36. Furthermore, alterations in gene expression were observed in the stim2 KO larvae, including a reduction in the expression of genes involved in light perception and an increase in the expression of anxa3a, a marker of activated microglia. Importantly, the thinning of RGCs and the IPL, changes in microglial function, and impaired vision in stim2 KO zebrafish resemble pathologies in glaucoma37,38,39.
The present study sought to understand the mechanisms that are responsible for vision problems that are caused by the loss of Stim2. The objective of this study was to ascertain whether Stim2 deficiency affects the gene expression profile in the retina or in a particular subset of neuronal cells, and whether the observed effects are associated with the vision deficits detected in stim2 KO larvae. We used bulk RNA sequencing (RNA-seq), single-cell RNA (scRNA)-seq, immunohistochemistry, confocal microscopy, and transmission electron microscopy to identify differences between stim2 KO zebrafish and a line that expressed both forms of Stim2 (referred to as control). We used zebrafish lines with genetically encoded fluorescent probe under a neuronal promoter to investigate the impact of stim2 KO in fluorescence-activated cell sorting (FACS)-purified neurons by scRNA-seq. We observed insulin among differentially expressed genes (DEGs) in specific neuronal cell clusters, which might underlie STIM2 associated pathologies at the cellular level. We found that features that are characteristic of glaucoma, such as reduction in IPL width and the number of ganglion cells and their dendrites, were also observed in stim2 KO zebrafish. Moreover, the analysis of γ-aminobutyric acid (GABA)-positive cells in the inner nuclear layer (INL) revealed their reduction, suggesting the importance of Stim2 in GABA-dependent neuronal properties.
The present study has demonstrated that Stim2 plays a pivotal role in the regulation of neuronal insulin expression, as well as in the maintenance of proper retinal morphology and function. Altogether, our results suggest that zebrafish depleted of Stim2 is a suitable model to investigate glaucoma-related changes.
Results
Bulk transcriptomics analyses in stim2 KO zebrafish
In samples from whole stim2 KO zebrafish larvae, vast changes in gene expression were observed36. However, it remained unknown whether Stim2 is also important in transcriptional regulation in the retina. To characterize changes in retinal gene expression patterns that are induced by the loss of Stim2, we performed bulk RNA-seq in micro-dissected eyes from 5-days postfertilization (dpf) stim2 KO larvae. We identified 32520 genes that were expressed across all samples, which was reduced to 22145 after filtering for low read count. Correlation analyses suggested no significant differences between genotypes (Fig. 1Sa). The top 20 most highly expressed genes, based on log-normalized count data, are shown in Fig. 1Sb and contained several marker genes for eyes (opn1-sw1 and cgrygm2d10, encoding opsin 1 and crystallin, respectively), confirming eye-origin of the analyzed samples. Differential gene expression analyses (padj [p-values adjusted] < 0.05) revealed four downregulated genes and one gene showing a tendency towards upregulation (padj = 0.06) (Table 1). However, the functions of protein products of those genes have not been elucidated and are based only on predictions, which makes it challenging to relate them with previously observed problems with vision and alterations of the stratum pigmentosum36.
Among the downregulated genes were si:dkey-159n16.2 (which encodes a protein that is predicted to have adenosine triphosphate [ATP] binding activity and involved in intracellular signal transduction), zgc:110699 (which is predicted to have guanosine triphosphate [GTP] binding activity and GTPase activity and involved in signal transduction), si:ch211-226h7.8 (which is predicted to be involved in neutrophil migration and active in the apical plasma membrane, cell surface, and extracellular space [orthologous to human glycoprotein]), and tss2.2 (which is predicted to have cysteine-type endopeptidase activity, involved in the immune response and proteolysis, and localized to the extracellular space and lysosome). The human ortholog of this gene (CTSS) encodes cathepsin S and is implicated in neuropathy. It is expressed in blood, macrophages, and the yolk syncytial layer. The only gene that was upregulated was si:ch211-214b16.4, which encodes a protein that is predicted to have ATP binding activity and involved in intracellular signal transduction.
In summary, the bulk RNA-seq analysis of the eye did not identify genes that could possibly explain vision impairment in stim2 KO zebrafish. However, the earlier bulk RNA-seq of whole stim2 KO larvae identified downregulated genes that are related to light perception36. This prompted us to search for genes in separated neuronal cells using scRNA-seq in the samples extracted from the brain of larval zebrafish rather than the eyes.
Graph-based clustering of single-cell transcriptome analysis reveals distinct neuronal cell populations
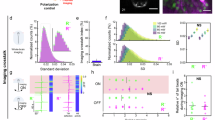
Single-cell RNA-seq was performed on brain cells of neuronal origin of 5 dpf zebrafish larvae that expressed green fluorescent probe under the huc/elavl3 promoter40 (Figs. 1a, 2S). Single-cell suspensions were obtained by the enzymatic and mechanical digestion41 of heads without eyes from two fish lines: stim2 KO and control. After obtaining the single-cell suspension, the cells were sorted by fluorescence-activated cell sorting (FACS) based on their green fluorescence (Fig. 3S).
Single-cell RNA-seq analysis of cells of neuronal origin. (a) Schematic of cell preparation and processing of the 5 dpf zebrafish brain. The procedure begins with the dissecting of the zebrafish larvae head and removal of the eyes. The larvae heads are then dissociated to create a single-cell suspension. Fluorescence-activated cell sorting (FACS) is used to isolate GCaMP5G-positive cells, which are indicative of neuronal origin. These cells are encapsulated in droplets using 10× Genomics technology for single-cell RNA sequencing (scRNA-seq). The encapsulated cells undergo library preparation, sequencing, and subsequent data analysis to assess gene expression. (b) UMAP representation demonstrates the distribution of control and stim2 KO zebrafish brain cells of neuronal origin. A total of 27 clusters were identified. Upright triangle symbols (Δ) show clusters with DEGs between stim2 KO and control cells (adjusted p < 0.05). Thirty larvae were used for each sample, and two samples of each line stim2 KO and controls were sequenced.
Twenty-seven distinct cell clusters were identified and visualized using the Uniform Manifold Approximation and Projection (UMAP) approach (Fig. 1b). Based on known marker genes that were provided by the Daniocell database42, we assigned cell types to clusters. Twenty-three clusters were defined in this way, but remaining four were devoid of any known cell types. The top 10 gene markers for each cluster are shown in Table 1S.
Differentially expressed genes in clusters of stim2 KO zebrafish are associated with the GABA-ergic neurons
Differentially expressed genes in clusters across conditions were identified with the nonparametric Wilcoxon rank sum test. Differentially expressed genes with adjusted p-values below 0.05 and a minimum two-fold change were found in 10 clusters. Interestingly, more than half (6/10) of the identified clusters with DEGs contain GABAergic neurons, including GABAergic retinal interneurons (cluster 2Δ) and amacrine retinal interneurons (cluster 3Δ; Table 2; Fig. 2a shows volcano plots for these two clusters). In GABAergic retinal interneurons (cluster 2Δ), two genes were upregulated: cart4 (eight-fold) and CR848784.2 (five-fold). First one encodes cocaine- and amphetamine-regulated transcript 4, the latter is unannotated. In the same cluster, two genes were downregulated: si:dkey-22i16.4 (52-fold) and CR626886.1 (35-fold). In amacrine retinal interneurons (cluster 3Δ), there were two downregulated genes: ins (38-fold) and si:dkey-22i16.4 (935-fold). Overall, ins, which encodes insulin, was downregulated at least 38-fold in six clusters, and si:dkey-22i16.4, which encodes a secretory calcium-binding phosphoprotein, was downregulated at least 20-fold in four clusters (Table 2, Fig. 4S). CR848784.2 was upregulated in three clusters at least five-fold, and acyp1 was upregulated in two clusters at least three-fold (Table 2). The latter encodes a small cytosolic enzyme that catalyzes hydrolysis of the carboxyl-phosphate bond of acylphosphates and is widely expressed in tissues, including the retina.
Differentially expressed genes in different clusters in stim2 KO zebrafish and the GO analysis. (a) Volcano plots of differentially expressed genes in clusters 2Δ and 3Δ. Genes significantly deregulated in each cluster are labeled. Horizontal dotted line indicates p-value adjusted = 0.05; green dot represent genes with not-significantly changed expression; red dots represent genes with significantly changed expression (p-value adjusted < 0.05 and log2 fold change (FC) different from 0). (b) Gene Ontology enrichment analysis of DEGs showing molecular function (red bars), cellular component (orange bars), and biological process (blue bars) aspects of 10 clusters with DEGs. All GO terms are presented for p < 0.05 (Fisher’s test). Up to five GO terms are presented for each cluster. Δ, clusters with dysregulated genes identified; α, clusters with dysregulated proportion of cells.
To ascribe functional relevance to the DEGs, we performed GO enrichment analysis in the clusters that are shown in Table 2 in all three GO classes: biological process, cellular component, and molecular function (Fig. 2b). In most clusters with DEGs, negative regulation of feeding behavior (clusters 0Δ, 1Δ, 2Δ, 3Δ, 5Δ, 9Δ), positive regulation of pancreatic A cell differentiation (clusters 0Δ, 1Δ, 3Δ, 5Δ), and translation and cytoplasmic translation (clusters 7Δ, 10Δ, 14Δ) were affected (Fig. 2b). Interestingly, the dysregulated expression of genes in 5 dpf larvae that are related to the regulation of feeding behavior was accompanied by a substantial tendency toward a smaller weight in adult stim2 KO zebrafish (Fig. 3). Additionally, the observed changes in cell proportions in specific clusters, such as the increase in GABAergic retinal interneurons and amacrine retinal interneurons, underscore the potential impact of Stim2 on neuronal cell type composition and function.
Changes in weight between stim2 KO zebrafish and control adult fish. stim2 KO zebrafish weighed considerably less than controls. n = 22 stim2 KO (7 females, 15 males), 15 control (7 females, 8 males). The scatter dot plots represent mean values ± standard error of the mean. Statistical analyses were performed using an unpaired t-test with Welch’s correction. Data passed the Shapiro–Wilk normality test and Rout method for eliminating outliers.
Changes in the proportion of GABAergic type neurons between stim2 KO zebrafish and control
Proportions of cells were significantly affected (False Discovery Rate [FDR] < 0.05) in eight clusters. Except cluster 23, all others belonged to clusters with DEGs (Table 2, Fig. 4). Therefore, the majority of affected cell clusters were those containing GABAergic cells. In three clusters, the proportion of cells from stim2 KO larvae was elevated (by 15% in cluster 2Δ, by 12% in cluster 3Δ, and by 15% in cluster 5Δ; Table 2, Fig. 4). In five clusters from stim2 KO zebrafish, the number of cells decreased compared with control larvae (by 40% in cluster 7Δ, by 42% in cluster 10Δ, by 20% in cluster 14Δ, by 34% in cluster 17Δ, and by 38% in cluster 23 [neurons/blood?]). This analysis demonstrated that the loss of Stim2 not only affects gene expression in GABAergic neurons, but also has an impact on the number of those cells in the zebrafish larvae brain. The changes in cell proportions, particularly the increased number of GABAergic retinal interneurons and amacrine retinal interneurons in stim2 KO zebrafish, reflect a possible compensatory mechanism or a disruption in neuronal development and differentiation. This shift in cellular composition may contribute to altered synaptic connectivity and neurotransmission, potentially leading to functional impairments.
Cell proportions in stim2 KO zebrafish (white bars) and control (gray bars) clusters. Asterisks mark clusters with significant changes in cell proportions in stim2 KO relative to control. Significance of change in cells proportions in clusters were calculated using t-test with Benjamini and Hochberg FDRs (False Discover Rates, *FDR < 0.05, **FDR < 0.01, ***FDR < 0.001). The proportion of cells was calculated using the propeller function (speckle v. 1.2 package) with arcsin square root transformation (see “Materials and methods”).
Detection of GABA-positive cells and photoreceptors in stim2 KO and control zebrafish
Single-cell RNA sequencing of samples extracted from the brains of larval zebrafish revealed an increased proportion of cells expressing GABAergic amacrine interneuron markers in stim2 KO versus control (Table 2, Fig. 4). This is in contrast to previously observed thinner ganglion cell layer (GCL), which contains GABAergic amacrine neurons43, in the retina of adult stim2 KO zebrafish36. To elucidate the apparent discrepancy between these observations, we conducted an analysis of the number of amacrine cells in the GCL and inner nuclear layer (INL) of the larval retina. To identify amacrine cells that are present in the GCL and INL, anti-GABA antibodies were used. Additionally, to stain photoreceptors, anti-opsin antibodies were used (Fig. 5).
Immunofluorescence visualization of cells in the retina of 5 dpf zebrafish larvae. (a) Photoreceptors (orange arrows) were stained with anti-opsin antibodies (green), and amacrine cells (yellow arrows) were stained with GABA antibodies (red). Nuclei were stained with Hoechst 33342 (blue). (b) (left) Representative retina image stained as above. (middle) Masks covering the inner nuclear (INL) and photoreceptor layers (PL) (right). Labeled nuclei contained in those layers. (c) Cell densities among the investigated retina layers, estimated as the number of cells that were positive for opsin or GABA above the fluorescence and size thresholds and normalized to surface volume of the mask. The photoreceptor layer and INL were 1.29 × and 1.78 × larger in control retinas of 5 dpf larvae. n = 7 stim2 KO, 5 control. The scatter dot plots represent mean values ± standard error of the mean. In the case of the photoreceptor cell density analysis, the Mann–Whitney test was conducted because the data did not pass the Shapiro–Wilk normality test. Statistical analyses of INL cell density were performed using an unpaired t-test with Welch’s correction because the data passed the Shapiro–Wilk normality test. The Rout method for eliminating outliers did not reveal any outliers among the tested conditions.
The analysis of cell density in the INL, which consists of horizontal cell bodies, bipolar cells, and amacrine cells, showed a significant decrease (1.78 × lower) in the number of GABAergic neurons in the retina in stim2 KO, suggesting that Stim2 may be involved in the regulation of GABAergic synaptic transmission, which plays an important role in shaping the retinal response to visual stimuli44 (Fig. 5c). Quantitative analysis of the photoreceptor layer in stim2 KO zebrafish revealed a significant reduction in the density of opsin-positive cells, specifically those expressing rhodopsin, indicating a decrease in the rod photoreceptor population (Fig. 6c). Both results suggest that Stim2 plays an important role in rod-dependent vision, such as dark and light adaptation45, and explain our previously demonstrated impairment in light preference36.
Transmission electron microscopy analysis revealed a decrease in IPL width as a consequence of ganglion cell perturbations. Left images show control samples and right images—stim2 KO samples. (a) Narrowing of the IPL in stim2 KO retina with visible malformations of ganglion cell dendrites. The yellow dotted lines indicate borders of the IPL. (b) The number of dendrites decreased in the IPL in stim2 KO zebrafish, and their shape was altered. Arrows point to dendrites. (c) Narrowing of the GCL in stim2 KO zebrafish, with a substantial decrease in the number of ganglion cells. Yellow dotted lines indicate borders of the GCL. The scatter dot plots represent mean values ± standard error of the mean. Images from at least four larvae were analyzed in (a–c). Statistical analyses were performed using an unpaired t-test with Welch’s correction. Data passed the Shapiro–Wilk normality test. (d) The number of microglia within GCL increased (arrows indicate the microglia cells). The Mann–Whitney test was conducted because the data did not pass the Shapiro–Wilk normality test; at least 5 larvae per variant were analyzed.
Transmission electron microscopy analysis reveal malformation in the stim2 KO retina structure
To comprehensively assess the impact of Stim2 deficiency on retinal ultrastructure, we conducted a thorough transmission electron microscopy (TEM) analysis across all retinal layers, with particular focus on the inner plexiform layer (IPL), ganglion cell layer (GCL), and photoreceptor layer. This systematic examination allowed us to identify and characterize structural alterations associated with the stim2 KO. In 5 dpf stim2 KO larvae, the GCL, INL, and IPL (also the inner synaptic layer) were narrowed compared with controls36. The IPL contains synapses between axons of bipolar cells and dendrites of ganglion cells. A decrease in the width of the IPL (Fig. 6a, c) and a reduction of the number of dendrites, suggesting their retraction, were also observed (Fig. 6a, arrows, 6c). Moreover, TEM analysis revealed that the number of ganglion cells was lower in stim2 KO zebrafish (Fig. 6b, c). Additionally a substantial increase in the number of microglia cells was evident (Fig. 6d), which is consistent with our previously published data suggesting their increased activation36. Altogether, our results indicate for the first time that lack of Stim2 might be involved in shaping GABAergic connectivity in the retina likely by the modulation of microglia activation.
Further quantitative analysis of photoreceptor mitochondria ultrastructure revealed significant alterations in stim2 KO zebrafish (Fig. 7). Specifically, the cristae area in photoreceptor cells of stim2 KO zebrafish was markedly reduced compared to control retinas (Fig. 7). This reduction in cristae area suggests a potential impact of Stim2 deficiency on mitochondrial morphology. Given the critical role of mitochondrial cristae in energy production, these structural changes may have profound implications for energy metabolism in retinal photoreceptors. This finding provides insight into the potential mechanisms by which Stim2 deficiency could affect photoreceptor function and survival, potentially contributing to the observed visual deficits in our model.
Cristae area in mitochondria of zebrafish photoreceptors. (a) Transmission electron microscopy image of mitochondria from 5 dpf zebrafish larvae with hand-labeled cristae. (b) Ratio of cristae area to area of mitochondria measured using ImageJ software in three pairs of control and stim2 KO zebrafish. Each pair contained at least 40 TEM images of 190 stim2 KO mitochondria and 160 controls. The data were analyzed using unpaired t-test.
Discussion
Glaucoma comprises a group of neurodegenerative diseases of optic nerve that can be caused by different factors and has a complex etiology. During glaucoma, RGCs and their axons undergo numerous alterations, including interruption of the retrograde axonal transport of neurotrophic factors, oxidative stress, ER stress, N-methyl-D-aspartate (NMDA) receptor activation, and intracellular Ca2+ overload6. Damage to RGC axons results in their progressive neurodegeneration and vision loss3. Axonal degeneration triggers rapid alterations of RGC dendritic arbors, leading to synaptic rearrangements (reviewed by46). Retinal ganglion cell dendrites receive synaptic inputs from bipolar and amacrine cells, allowing the flow of visual information. In glaucoma, the function of RGCs may be compromised due to alterations in their dendritic connections with bipolar cells within the IPL.
We propose that the stim2 KO zebrafish might be useful for studying some features of glaucoma, such as the thinning of RGCs and the IPL, which we observed in stim2 KO larvae36 and occur in glaucoma37,38,39. As Stim2 KO mice exhibited pronounced cognitive deficits and survived only a few weeks after birth47,48, therefore investigating long-term effects of Stim2 KO in rodents is not feasible, making zebrafish an attractive alternative. We generated zebrafish KO of stim2a, stim2b, and both, which were viable34,35. The lack of both Stim2 isoforms affected vision-related behaviors36 and retinal structure in stim2 KO zebrafish. The phototaxis of stim2 KO larvae was found to be disrupted, with the majority of the examined fish exhibiting no light preference or even a reversal of preference in comparison to wild-type fish36. A comparable, though less pronounced, phenomenon was observed in the case of single stim2a and stim2b KO34,35. Additionally, the visual-motor response of stim2 KO larvae was abnormal, as they did not increase their mobility upon switching the light off36, in contrast to wild-type zebrafish and single stim2a and stim2b KO34,35. These findings suggest that both zebrafish Stim2 isoforms may be involved in visual perception and that the effects of their depletion are cumulative. Moreover, stim2 KO larvae and adults possessed a thicker RPE36, which plays a key role in maintaining the photoreceptor layer49. Histological and TEM analyses further demonstrate the structural and cellular alterations in the retina of stim2 KO zebrafish. The reduced number of GABAergic neurons and photoreceptors, along with the increased microglial activation, indicate that Stim2 plays a crucial role in maintaining retinal integrity and function. Our previous work demonstrated that Stim2 deficiency leads to increased anxa3 expression, a marker of microglial activation30. This finding is particularly intriguing given the crucial role of microglia in maintaining neural homeostasis through their dynamic functions, including migration, phagocytosis, and cytokine production. Previous research has established a link between store-operated calcium entry (SOCE) and microglial properties, particularly in regulating phagocytosis and cytokine secretion50. The dendritic synapse loss observed in our study may be attributed to the disruption of Ca2⁺ homeostasis in microglia and their subsequent interactions with GABAergic neurons. Our current TEM data, showing an increased number of microglia in the ganglion cell layer (GCL), corroborates our earlier findings36.
STIM2 plays a significant role in regulating gene expression in neurons through several mechanisms. One of these is its involvement in SOCE, which can activate transcription factors such as CREB, NFAT and NFκB and thus modulate gene expression51. Another is the maintenance of basal calcium levels in neurons, which is crucial for normal neuronal excitability52,53. Although there is currently no direct evidence linking STIM1/2 with glaucoma, TRPC1 and TRPC6 channels, which are known interacting partners of STIM isoforms, were upregulated in glaucomatous lamina cribrosa leading to NFAT4-mediated gene expression remodeling54.
In both stim2a and stim2b single knockout zebrafish the changes in gene expression have been observed34,35. However, the analysis were conducted on samples from whole larvae. Consequently, it remains unclear whether the regulation of gene expression by Stim2 is dependent on the neuronal type, brain region or physiological context. This makes it challenging to identify DEGs specific to particular cell populations. To address this issue, we employed bulk RNA-seq analysis of the eye and scRNA-seq in neurons to examine alterations in gene expression in diverse neuronal cells following Stim2 depletion.
The bulk RNA-seq of eye samples revealed only a limited number of differentially expressed genes, which initially posed challenges in correlating molecular changes with the observed phenotype. This discrepancy might be attributed to the limitations of bulk analysis, where subtle alterations in specific cell subsets could be obscured by the overall tissue composition.
Interestingly, previously performed bulk RNA-seq analysis of whole larvae yielded more pronounced results. We observed downregulation of key adhesion molecules crucial for retinal structure maintenance, including pcdh15b, prph2b, rom1a, and rom1b. Additionally, the eye development transcription factor crx showed reduced expression. Notably, we detected a significant decrease in various vision-related genes, encompassing opsins, rhodopsin kinase (grk1a), phosphodiesterase (pde6ha), cGMP-gated cation channels, and metabotropic glutamate receptor 6 (grm6b)36.
To delve deeper into the neuronal impacts of Stim2 loss, we conducted single-cell RNA-seq on dissected zebrafish brains. This approach allowed us to examine gene expression profiles in distinct neuronal subpopulations. The contrast between the modest changes observed in eye samples and the more pronounced differences in whole larvae and brain samples is particularly striking. This disparity suggests that Stim2 may exert a stronger influence on gene expression in brain neurons compared to ocular tissues. In 10 out of 27 identified clusters of cells of neuronal origin from brains of 5 dpf stim2 KO zebrafish statistically significant differences in gene expression in comparison to control were detected. In three clusters (amacrine retinal interneurons, GABAergic retinal interneurons and GABAergic diencephalon cells), the proportion of cells increased in stim2 KO. Thus, there is an apparent discrepancy between the increased proportion of cells expressing GABAergic amacrine interneuron markers in the brain and reduced numbers of GABAergic amacrine cells in the retina of stim2 KO zebrafish. This modest yet statistically significant increase in the population of these cells may be indicative of a compensatory effect in response to the reduction in GABAergic amacrine cells in the retina. Additionally, it is important to consider that the observed alterations in the proportion of GABAergic cells in different regions of the brain may reflect aberrant development of these cells due to dysregulated gene expression. It is well-established that Ca2+ signaling plays a crucial role in the maturation and migration of GABAergic interneurons, suggesting that the absence of Stim2 could potentially impact this process55,56. Moreover, in five clusters with DEGs, the proportion of cells in stim2 KO zebrafish versus control larvae significantly decreased, including GABAergic diencephalon and GABAergic optic tectum cells. A disrupted balance between inhibitory and excitatory signaling in the optic tectum affects neuronal activity and may be linked with our previous observations of a higher frequency of Ca2+ oscillations in optic tectum neurons in single stim2a34 and stim2b35 KO lines.
In clusters with higher or unchanged numbers of cells (0Δ, 1Δ, 2Δ, 3Δ, 5Δ), the negative regulation of feeding behavior and, surprisingly, the positive regulation of pancreatic A cell differentiation were observed. The effect on feeding correlates well with the observed loss of weight in stim2 KO zebrafish and was likely attributable to downregulation of the ins (which encodes insulin). Moreover, it correlates with the lower deposition of glycogen in the liver in adult stim2 KO zebrafish36. These findings suggest a systemic impact of loss of Stim2 on metabolic regulation and energy homeostasis. Whether Stim2 participates in insulin metabolism in neurons has not yet been established, but our work indicates its role in the neuronal expression of ins gene as well as of si:dkey-22i16.4.
The si:dkey-22i16.4, a gene encoding a secretory calcium-binding phosphoprotein, has not been previously linked to vision. This protein is involved in calcification processes, such as tooth formation in medaka and zebrafish57,58. The si:dkey-22i16.4 is expressed in various tissues in cyprinid fish, including the eye and brain59. Whether and how the decrease in expression of si:dkey-22i16.4 in retinal cell clusters in stim2 KO zebrafish is linked to the observed glaucoma-like retinal pathology are still unclear. However, the fact that both the si:dkey-22i16.4 and ins genes are highly downregulated in the same cell clusters is unlikely accidental. Notably, the calcification of trabecular meshwork was observed in open-angle glaucoma60. Aortic calcifications and dental dysplasia were described in Singleton-Merten syndrome, a rare immunogenetic disorder that features juvenile open-angle glaucoma61,62. Calcification of the carotid artery into the optic canal may play a role in the pathogenesis of low-tension glaucoma, called soft glaucoma63 (reviewed by64). It can thus be postulated that the disruption of calcification associated with the downregulation of si:dkey-22i16.4 may underlie the glaucoma-like features observed in stim2 KO zebrafish.
Insulin may be implicated in the pathology of vision. Insulin promotes substantial dendrite and possibly synapse regeneration, mediated by mammalian/mechanistic target of rapamycin complex 1 (mTORC1) and mTORC265. Several studies demonstrated the potential of modulating insulin signaling in the treatment of neurodegenerative diseases, including glaucoma66. Diabetic patients have a higher risk of glaucoma67,68,69,70, whereby cataracts are formed earlier and progress faster71. Thus, our observation that ins expression significantly decreased in selected cell clusters, such as GABAergic retinal interneurons and amacrine retinal interneurons, supports links between glucose levels and eye pathology.
In this study, we present novel findings on the relationship between Stim2 and neuronal insulin expression. While previous research has established links between SOCE and insulin secretion in pancreatic β cells72, our data implies a unique role for Stim2 in neuronal ins gene transcription. We demonstrated that Stim2 deficiency leads to significantly reduced ins expression in several identified neuronal clusters (Fig. 4S), that include in the majority GABAergic neurons. This observation aligns with and extends prior work by Molnár et al.73, which identified GABAergic neurogliaform cells as local sources of insulin in the cerebral cortex. STIM2 role in the regulation of neuronal ins expression has not been previously reported. Given the critical role of insulin in neuronal survival, plasticity, and cognitive function74, these findings open an unexplored area for investigation of the interplay between calcium signaling, insulin expression, and GABAergic neurons function. Furthermore, these results provide a potential mechanistic link to studies demonstrating the effects of intranasal insulin on brain function, suggesting a complex regulatory network involving SOCE, Stim2, and neuronal insulin production75.This work provide a basis for further investigation into the tissue-specific regulation of ins expression and its implications for neurological function and disorders.
Retinal ganglion cells contain the highest number of mitochondria compared with other retinal cells to fulfill the higher energy requirements of efficient action potential propagation involved in highly dynamic visual-motor communication70. In glaucoma, RGC mitochondria undergo oxidative stress, leading to greater susceptibility to degeneration70. Similarly, photoreceptors also rely heavily on mitochondrial function for their high energy demands and calcium homeostasis76. In stim2 KO zebrafish, we observed significant loss of cristae in mitochondria of photoreceptors (Fig. 8), suggesting that STIM2 plays a crucial role in maintaining mitochondrial integrity in these cells. This observation aligns with previous studies demonstrating the importance of calcium signaling in photoreceptor function and survival77. However, the limited scope of this study does not allow for the identification of the exact mechanism of the observed changes. It is possible that loss of Stim2 disrupts mitochondrial Ca2+ homeostasis, which may result in cristae disorganization and an impairment of the energetic balance of retinal cells.
Pleiotrophic effect of stim2 knockout resembling glaucoma like phenotype in zebrafish. Loss of Stim2 leads to impairment of GABAergic connections, resulting in loss of ganglion cells, causing changes in light perception. Increase in microglia number and their activation36, glycogen deficiency36 as well as mitochondria alterations may be a consequence of the insulin downregulation seen in stim2 KO. GCL, Ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; PL, photoreceptor layer.
In conclusion, our study elucidates the multifaceted role of Stim2 in neuronal gene regulation and cellular physiology, offering insights into its involvement in shaping neuronal connectivity and potential implications for glaucoma pathophysiology. We observed significant structural and functional alterations in the retina stim2 KO zebrafish, including dendrite retraction in the inner plexiform layer (IPL), decreased dendrite numbers, substantial reduction in the retinal ganglion cell (RGC) layer. Furthermore, this study reveals a significant decrease in GABAergic neurons and a considerably smaller population of rhodopsin-positive rod cells in stim2 KO zebrafish. These results implicate Stim2 in the regulation of GABAergic synaptic transmission (Fig. 8), which is crucial for shaping retinal responses to visual stimuli44 and rod-dependent vision, including dark and light adaptation45. This observation aligns with our previous demonstration of light preference loss in stim2 KO larvae36. The specific changes in gene expression observed in neuronal cell clusters point to alterations in the insulin-mediated signaling (Fig. 8). These findings provide a basis for further investigation into the tissue-specific regulation of insulin expression and its implications for neurological function and disorders. The complex interplay between Stim2, calcium homeostasis, and insulin signaling in the context of microglial-GABAergic neuron communication presents a promising area for future investigation.
However, our observation that lack of Stim2 in zebrafish induces changes in retina similar to those observed in glaucoma have some limitations. The specific pathway responsible for the observed pathologies remains unknown, and it has not be determined if both of the genes that were found to be highly downregulated participate in this process. Future studies will address these issues and help to establish the suitability of stim2 KO zebrafish as a model of glaucoma-like pathology taking into account that any single model perfectly replicates human glaucoma.
Materials and methods
Statement of ethics
All experiments were approved by Advisory Team for Animal Welfare Committee of International Institute of Molecular and Cell Biology in Warsaw and conducted in accordance with the European Communities Council Directive (63/2010/EEC) as described in detail previously36. All zebrafish were maintained according to internal regulations in the Zebrafish Core Facility (PL14656251, registry of the District Veterinary Inspectorate in Warsaw; 064 and 051, registry of the Ministry of Science and Higher Education) at the International Institute of Molecular and Cell Biology (IIMCB) in Warsaw (IN-MOL-CELL infrastructure). This study is reported in accordance with ARRIVE guidelines78.
Animal husbandry
The Tg(HuC:GCaMP5G) zebrafish line79, AB zebrafish line, and stim2 KO line that expressed GCaMP5G under the same promoter were used. The animals were maintained in the Zebrafish Core Facility under standard conditions80.
Bulk RNA-seq
The procedure for bulk RNA-seq analysis was conducted according to our previous study36. Total RNA was isolated from zebrafish eyes. To synthesize sequencing libraries, a two-step approach was applied. Total RNA was subjected to mRNA enrichment using the NEBNext Poly(A) mRNA Magnetic Isolation Module (E7490, NEB), followed by the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (E7760, NEB) according to the manufacturer’s guidelines with minor adjustments. In brief, 1 μg total RNA was used to enrich the polyadenylated RNA fraction. The obtained RNA was fragmented for 12 min at 94 °C to target the insert size around 400 bp, followed by extended first- and second-strand synthesis. To enrich adaptor-ligated molecules, samples were pre-amplified with four cycles and left aside on ice. To determine an optimal number of polymerase chain reaction (PCR) cycles and limit the possibility of generating PCR duplicates and artifacts, a subsequent quantitative PCR reaction on previously pre-amplified samples was performed. Final libraries were validated in terms of quality and quantity by a Quantus fluorometer (Promega) and Tapestation 2020 High Sensitivity D1000 assay (Agilent Technologies), respectively. Pair-end sequencing (2 × 150 bp) was performed on an Illumina NovaSeq 6000 using a NovaSeq 6000 S4 flow cell (Illumina) to target a depth of 25–30 million reads per sample. The experimental setup involved three replicates per condition (stim2 KO vs. control), consisting of 35 eyes per repetition.
The FastQC tool (v. 0.11.8) was used to assess quality of the obtained raw RNA-seq reads, followed by read mapping to the zebrafish reference genome (GRCz11 v. 109) using STAR aligner (v. 2.7)81, resulting in a mappability rate of 82–87%. Sequencing reads were further analyzed in the R programming language (v. 4.0.2), and DEGs were identified by the DESeq2 package82 according to the default workflow. Principal component analysis was performed on normalized read counts that were transformed to the log2 scale by the plotPCA function from the same package. The ggplot2 package was used for plot generation.
Single-cell RNA-seq
Isolation of single-cell suspension
Larvae at 5 dpf were anesthetized by tricaine methanesulfonate MS222 (cat. no. E10521, Sigma-Adlrich) and heads were dissected in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (cat. no 21041025, Gibco) with 10% fetal bovine serum (Gibco). Eyes were surgically removed from the zebrafish to focus on cells of neuronal origin. Single-cell suspensions were obtained by the enzymatic and mechanical digestion41 of heads without eyes that were isolated from two zebrafish lines (stim2 KO and control).
Isolation of cells of neuronal origin
Immediately after the isolation of single-cell suspensions, the cells were sorted based on GCaMP5G fluorescence (488 nm excitation wavelength, 510 nm emission wavelength) to obtain cells of neuronal origin. GCaMP5 was expressed under the elav/Huc promoter, which is an early marker of neuronal cells40,83. Cell sorting was performed with a BD FACSAria II (BD Biosciences, Franklin Lakes, NJ, USA) with support from the Core Facility at IIMCB in Warsaw (Fig. 3S). Cell viability was measured using the trypan blue staining method. When the viability of cells was ~ 80%, the cells were immediately loaded on the 10× Chromium system for droplet encapsulation.
Droplet encapsulation, library preparation, and sequencing
Approximately 8000 cells were sorted and suspended in a ~ 40 μl volume. Cells were loaded according to the Chromium single cell 3′ kit’s standard protocol (V3 chemistry). To prepare the cells for droplet-based sequencing, GCaMP5G-positive single-cell suspensions were carefully mixed with reverse transcription mix before loading the cells on the 10X Genomics Chromium system according to the standard manufacturer’s protocol. During encapsulation, the cells were lysed within the droplet, and they released polyadenylated RNA bound to the barcoded bead, which was encapsulated with the cell. Following the 10X Genomics user manual’s guidelines, the droplets were directly subjected to reverse transcription, the emulsion was broken, and cDNA was purified using Silane beads. After the amplification of cDNA with 13 cycles, purification and quantification were performed. The 10× Genomics single-cell RNA-seq library preparation (involving fragmentation, dA-tailing, adapter ligation, and 12-cycle indexing PCR) was performed. After quantification, the libraries were sequenced at the Nencki Institute of Experimental Biology on an Illumina NextSeq 550 machine using a HighOutput flow cell in paired-end mode (R1: 26 cycles; I1: 8 cycles; R2: 57 cycles), thus generating 80–125 million fragments.
scRNA-seq data analysis and identification of cell clusters
We obtained primary assembly of the zebrafish genome (Danio rerio GRCz11) and transcriptome annotation files from ensembl (release 105). Raw reads from fastq files were mapped to the aforementioned zebrafish reference genome, and gene expression was quantified with the STARSolo mode of STAR84 for each sample with the following parameters: --soloType CB_UMI_Simple, --soloCBwhitelist 3M-february-2018.txt (from Cell Ranger), --soloUMIlen 12, --soloCBmatchWLtype 1MM_multi_Nbase_pseudocounts, --soloUMIfiltering MultiGeneUMI_CR, --soloUMIdedup 1MM_CR, --soloCellFilter EmptyDrops_CR). In the preprocessing step that was conducted with the Seurat package (v. 5.0.1)85, we filtered out genes that were not expressed in at least three cells and kept only cells that expressed at least 100 genes and contained less than 10% mitochondrial genes. Additionally, to avoid doublets, cells with more than 2500 genes were removed. After this filtering step, 22889 cells were kept in both control and stim2 KO samples with 20968 features identified. We then integrated all datasets from four samples (two replicates each for transgenic/control and stim2 KO conditions) with the harmony package (v. 1.2)86.
The neighborhood graph was embedded using UMAP, and Leiden clustering (resolution = 1) was performed. The FindAllMarkers, FindConservedMarkers, and FindMarkers functions of Seurat were used to identify marker genes per cluster and differential gene expression in general between stim2 KO and control for each cluster, as well as for all clusters per the entire dataset. We utilized curated gene expression data from the Daniocell database (daniocell.nichd.nih.gov) for all cell types in zebrafish neural tissues and eye neuronal cells and compiled a list of marker genes with the highly expressed genes in each cell type. The nomenclature used for the cell clusters is based on common gene markers associated with specific cell types. Consequently, some cluster names may appear to refer to retinal cell types, such as “retinal interneurons”, despite analysis of samples obtained from the brain. This phenomenon is attributable to the overlap of specific gene expression patterns between brain and retinal cell types, reflecting their shared developmental origins and functional similarities, thus these clusters represent brain cells expressing markers similar to those found in retinal cell types, rather than actual retinal cells.
The proportion of cells was calculated using the propeller function with arcsin square root transformation of proportions from the speckle v. 1.2 package87. Plots of proportions of cells were generated with GraphPad Prism software. Gene Ontology terms were identified with the topGO v. 2.54 package for DEGs or for marker genes in each cluster88. All steps that required the R programming language were performed in R v. 4.3.1.
Gene Ontology
To identify functionally related genes the elim method from the topGO v. 2.54.0 and the org.Dr.eg.db v. 3.18.0 R packages was used88. Two gene sets were used to identified GO terms enrichment. First gene set were selected from marker genes for clusters (identified by the FindAllMarkers function from the Seurat package; p-value adjusted less than 0.05). Second gene set was selected from differentially expressed genes (DEGs) in each cluster (identified by the FindMarkers function from the Seurat package; p-value adjusted less than 0.05). In both cases gene identifiers with p-value adjusted greater than 0.05 (calculated with FindAllMarkers or FindMarkers functions from the Seurat package) were used as background genes. The statistical significance for GO terms enrichment was calculated with Fisher’s exact test.
Transmission electron microscopy of zebrafish larvae
Zebrafish larvae at 5 dpf were euthanized using Tricane. The animals were fixed in 2.5% glutaraldehyde for 24 h at 4 °C, washed in phosphate-buffered saline (PBS), and postfixed with 1% osmium tetroxide for 1 h, followed by washing with water and staining with 1% aqueous uranyl acetate overnight at 4 °C. Next, the larvae were dehydrated with increasing concentrations of ethanol at room temperature and infiltrated with epoxy resin (Sigma Aldrich, St. Louis, MO, USA, catalog no. 45-359-1EA-F). Samples were then taken for polymerization for 48 h at 60 ºC. Polymerized blocks were trimmed with tissue processor (Leica EM TP) and cut with an ultramicrotome (EM UC7, Leica) to make ultrathin (70 nm thick) sections, which were collected on nickel grids (200 mesh, Agar Scientific, catalog no. G2200N). Specimen grids were examined with a Tecnai T12 BioTwin transmission electron microscope (FEI, Hillsboro, OR, USA) that was equipped with a 16 megapixel TemCam-F416 camera (TVIPS GmbH) in the Microscopy and Cytometry Facility at IIMCB in Warsaw.
Electron microscopy analysis of retina layers
The retina is a very well-organized structure. It is characterized by subsequent layers of cells and their processes. This enables easy identification of particular layers. In our analysis, we measured widths of the IPL and GCL, and we quantified IPL dendrites and GCL neurons. Widths of the IPL and GCL were manually measured with ImageJ software on a single low-magnification (1200 ×) image where the entire surface of the layer was visible. The number of dendrites was assessed from images that were obtained at a larger magnification (4800 ×). Individual images were composed in CorelDRAW to show the surface visible in the low-magnification image. The image prepared in this way was exported to a tiff bitmap. We identified IPL dendrites as an object with a single membrane and eventually with mitochondria and without the signs of microtubule bundles and vesicles. These last two features enable us to differentiate them from axons also present in this layer. Neurons of GCL were assessed on the base of the number of nuclei present in this layer and using images taken at a magnification of 1200x. All measurements were performed manually in ImageJ software. The obtained data were analyzed by GraphPad Prism 9.0 software using an unpaired t-test with Welch’s correction. For each genetic variant, at least four samples were analyzed. To test the normality of the data, the Shapiro–Wilk test was performed.
For measurements of mitochondria in photoreceptors, the ratio of cristae area to the area of mitochondria was calculated using ImageJ software in the three pairs of control and stim2 KO zebrafish according to a previous report89. Each pair contained at least 40 electron microscopy images of mitochondria. The total number of mitochondria that were analyzed was 190 in stim2 KO zebrafish and 160 in controls. The data were analyzed using an unpaired t-test.
Visualization of retinal cells and counting
Zebrafish larvae at 5 dpf were euthanized using Tricane, fixed in 4% paraformaldehyde overnight, incubated in 30% sucrose overnight, transferred to optimal cutting temperature compound in liquid nitrogen, and cut into 20 µm sections using a Leica cryostat. Sections were dried overnight, incubated for 5 min at room temperature in PBS/1%Tween/0.1% dimethylsulfoxide, hydrated in PBS for 10–15 min, permeabilized for 10 min at RT in PBS/0.2% Triton X-100, blocked in 5% bovine serum albumin with Triton-X100 for 30 min at room temperature in a humid chamber, incubated with primary antibodies in 5% bovine serum albumin (for photoreceptors: monoclonal anti-opsin, catalog no. O4886, Sigma-Aldrich; for amacrine cells: anti-GABA, catalog no. 0000177418, Sigma-Aldrich) at room temperature overnight in a humid chamber, rinsed, and incubated with secondary antibody (for photoreceptors: Alexa Fluor 488 donkey anti-mouse IgG [H + L], lot no. 2147618, Invitrogen; for amacrine cells: Alexa Fluor 594 goat anti-rabbit IgG [H + L], lot no. WH322204, Invitrogen) in 5% bovine serum albumin at room temperature for 2 h in the dark in a humid chamber. Samples were washed and incubated for 15 min with Hoechst 33342 (Thermo Fisher; 1:1000 in PBS) at room temperature. After drying, mounting medium (Prolong Gold, catalog no. P36930, Invitrogen) was added to the slides. Z-stacks of images were acquired by confocal microscopy (Zeiss LSM 800; 40 × /1.3 oil objective).
To calculate cell densities of GABA-positive cells in the INL and opsin-positive cells in the outer nuclear layer, we applied a segmentation workflow in Imaris, Cellpose90, and ImageJ software. First, surfaces for both layers were drawn manually on three-dimensional image stacks in Imaris and converted to masks. Second, Hoechst-stained nuclei in the retina were three-dimensionally segmented using the “denoise” image restoration and cyto3 models in Cellpose90. Third, nuclear labels were thresholded for fluorescence signal, masked for each layer, thresholded for volume, and counted in ImageJ. To obtain cell densities, the counted nuclei were divided by mask volume. Because stim2 KO retinas had substantially lower GABA signals than controls, the threshold for GABA fluorescence in the above workflow was calculated automatically with the “Adjust Threshold” tool in ImageJ (default setting) to retain GABA-positive structures that were comparable in shape and volume.
Data availability
All data generated or analyzed during this study are included in this article (and its ementary Information files). RNA sequencing data (bulkRNAseq, scRNAseq) are available in the GEO repository under the accession number GSE264312 .
References
Wojda, U., Salinska, E. & Kuznicki, J. Calcium ions in neuronal degeneration. IUBMB Life 60, 575–590. https://doi.org/10.1002/iub.91 (2008).
Crish, S. D. & Calkins, D. J. Neurodegeneration in glaucoma: Progression and calcium-dependent intracellular mechanisms. Neuroscience 176, 1–11. https://doi.org/10.1016/j.neuroscience.2010.12.036 (2011).
Wang, A. Y. et al. Potential mechanisms of retinal ganglion cell type-specific vulnerability in glaucoma. Clin. Exp. Optom. 103, 562–571. https://doi.org/10.1111/cxo.13031 (2020).
Irnaten, M. & O’Brien, C. J. Calcium-signalling in human glaucoma lamina cribrosa myofibroblasts. Int. J. Mol. Sci. 2023, 24. https://doi.org/10.3390/ijms24021287 (2023).
Santos-Ledo, A., Pérez-Montes, C., DeOliveira-Mello, L., Arévalo, R. & Velasco, A. Oligodendrocyte origin and development in the zebrafish visual system. J. Comp. Neurol. 531, 515–527. https://doi.org/10.1002/cne.25440 (2023).
Munemasa, Y. & Kitaoka, Y. Molecular mechanisms of retinal ganglion cell degeneration in glaucoma and future prospects for cell body and axonal protection. Front. Cell Neurosci. 6, 60. https://doi.org/10.3389/fncel.2012.00060 (2012).
Vernazza, S., Oddone, F., Tirendi, S. & Bassi, A. M. Risk factors for retinal ganglion cell distress in glaucoma and neuroprotective potential intervention. Int. J. Mol. Sci. 2021, 22. https://doi.org/10.3390/ijms22157994 (2021).
McKernan, D. P., Guerin, M. B., O’Brien, C. J. & Cotter, T. G. A key role for calpains in retinal ganglion cell death. Invest. Ophthalmol. Vis. Sci. 48, 5420–5430. https://doi.org/10.1167/iovs.07-0287 (2007).
Huang, W., Fileta, J., Rawe, I., Qu, J. & Grosskreutz, C. L. Calpain activation in experimental glaucoma. Invest. Ophthalmol. Vis. Sci. 51, 3049–3054. https://doi.org/10.1167/iovs.09-4364 (2010).
Qu, J., Wang, D. & Grosskreutz, C. L. Mechanisms of retinal ganglion cell injury and defense in glaucoma. Exp. Eye Res. 91, 48–53. https://doi.org/10.1016/j.exer.2010.04.002 (2010).
Wang, Y. et al. Calpain-1 and calpain-2 play opposite roles in retinal ganglion cell degeneration induced by retinal ischemia/reperfusion injury. Neurobiol. Dis. 93, 121–128. https://doi.org/10.1016/j.nbd.2016.05.007 (2016).
Ryu, M. et al. Critical role of calpain in axonal damage-induced retinal ganglion cell death. J. Neurosci. Res. 90, 802–815. https://doi.org/10.1002/jnr.22800 (2012).
Osborne, N. N. et al. Effectiveness of levobetaxolol and timolol at blunting retinal ischaemia is related to their calcium and sodium blocking activities: Relevance to glaucoma. Brain Res Bull. 62, 525–528. https://doi.org/10.1016/s0361-9230(03)00070-4 (2004).
Chidlow, G., Wood, J. P. & Casson, R. J. Pharmacological neuroprotection for glaucoma. Drugs 67, 725–759. https://doi.org/10.2165/00003495-200767050-00006 (2007).
Boccaccini, A. et al. Novel frontiers in neuroprotective therapies in glaucoma: Molecular and clinical aspects. Mol. Aspects Med. 94, 101225. https://doi.org/10.1016/j.mam.2023.101225 (2023).
Guarino, B. D., Paruchuri, S. & Thodeti, C. K. The role of TRPV4 channels in ocular function and pathologies. Exp. Eye Res. 201, 108257. https://doi.org/10.1016/j.exer.2020.108257 (2020).
Cheung, L. T. Y., Manthey, A. L., Lai, J. S. M. & Chiu, K. Targeted delivery of mitochondrial calcium channel regulators: The future of glaucoma treatment?. Front. Neurosci. 11, 648. https://doi.org/10.3389/fnins.2017.00648 (2017).
Liu, H., Liu, H. & Prokosch, V. The relationship between mitochondria and neurodegeration in the eye: A review. Appl. Sci. 11, 7385 (2021).
Stavropoulos D et al. The role of mitophagy in glaucomatous neurodegeneration. Cells 2023, 12. https://doi.org/10.3390/cells12151969 (2023).
Molnar, T. et al. Store-operated channels regulate intracellular calcium in mammalian rods. J. Physiol. 590, 3465–3481. https://doi.org/10.1113/jphysiol.2012.234641 (2012).
Majewski, L. & Kuznicki, J. SOCE in neurons: Signaling or just refilling?. Biochim. Biophys. Acta 1853, 1940–1952. https://doi.org/10.1016/j.bbamcr.2015.01.019 (2015).
Wegierski, T. & Kuznicki, J. Neuronal calcium signaling via store-operated channels in health and disease. Cell Calcium 74, 102–111. https://doi.org/10.1016/j.ceca.2018.07.001 (2018).
Skopin, A. Y. et al. A novel modulator of STIM2-dependent store-operated Ca2+ channel activity. Acta Naturae 13, 140–146. https://doi.org/10.32607/actanaturae.11269 (2021).
Klejman, M. E. et al. Expression of STIM1 in brain and puncta-like co-localization of STIM1 and ORAI1 upon depletion of Ca(2+) store in neurons. Neurochem. Int. 54, 49–55. https://doi.org/10.1016/j.neuint.2008.10.005 (2009).
Gruszczynska-Biegala, J. & Kuznicki, J. Native STIM2 and ORAI1 proteins form a calcium-sensitive and thapsigargin-insensitive complex in cortical neurons. J. Neurochem. 126, 727–738. https://doi.org/10.1111/jnc.12320 (2013).
Gruszczynska-Biegala, J., Pomorski, P., Wisniewska, M. B. & Kuznicki, J. Differential roles for STIM1 and STIM2 in store-operated calcium entry in rat neurons. PLoS One 6, e19285. https://doi.org/10.1371/journal.pone.0019285 (2011).
http://www.informatics.jax.org/marker/phenotypes/MGI:2151156 (2023).
Zhai, Y. et al. Targeted exome sequencing of congenital cataracts related genes: Broadening the mutation spectrum and genotype-phenotype correlations in 27 Chinese Han families. Sci. Rep. 7, 1219. https://doi.org/10.1038/s41598-017-01182-9 (2017).
Rhodes, J. D. & Sanderson, J. The mechanisms of calcium homeostasis and signalling in the lens. Exp. Eye Res. 88, 226–234. https://doi.org/10.1016/j.exer.2008.10.025 (2009).
Gomez, N. M., Tamm, E. R. & Straubeta, O. Role of bestrophin-1 in store-operated calcium entry in retinal pigment epithelium. Pflugers Arch. 465, 481–495. https://doi.org/10.1007/s00424-012-1181-0 (2013).
Yang, I. H. et al. Involvement of STIM1 and Orai1 in EGF-mediated cell growth in retinal pigment epithelial cells. J. Biomed. Sci. 20, 41. https://doi.org/10.1186/1423-0127-20-41 (2013).
Chanaday, N. L. et al. Presynaptic store-operated Ca(2+) entry drives excitatory spontaneous neurotransmission and augments endoplasmic reticulum stress. Neuron 109, 1314-1332.e5. https://doi.org/10.1016/j.neuron.2021.02.023 (2021).
Wasilewska, I., Gupta, R. K., Palchevska, O. & Kuznicki, J. Identification of zebrafish calcium toolkit genes and their expression in the brain. Genes (Basel) 2019, 10. https://doi.org/10.3390/genes10030230 (2019).
Gupta, R. K., Wasilewska, I., Palchevska, O. & Kuznicki, J. Knockout of stim2a increases calcium oscillations in neurons and induces hyperactive-like phenotype in zebrafish larvae. Int. J. Mol. Sci. 2020, 21. https://doi.org/10.3390/ijms21176198 (2020).
Wasilewska, I., Gupta, R. K., Wojtas, B., Palchevska, O. & Kuznicki, J. stim2b Knockout induces hyperactivity and susceptibility to seizures in zebrafish larvae. Cells 2020, 9. https://doi.org/10.3390/cells9051285 (2020).
Wasilewska, I. et al. Lack of Stim2 affects vision-dependent behavior and sensitivity to hypoxia. Zebrafish 20, 146–159. https://doi.org/10.1089/zeb.2022.0068 (2023).
Boia, R. et al. Neuroprotective strategies for retinal ganglion cell degeneration: Current status and challenges ahead. Int. J. Mol. Sci. 2020, 21. https://doi.org/10.3390/ijms21072262 (2020).
Ahmad, I. & Subramani, M. Microglia: Friends or foes in glaucoma? A developmental perspective. Stem Cells Transl. Med. 11, 1210–1218. https://doi.org/10.1093/stcltm/szac077 (2022).
Tribble, J. R. et al. Neuroprotection in glaucoma: Mechanisms beyond intraocular pressure lowering. Mol. Aspects Med. 92, 101193. https://doi.org/10.1016/j.mam.2023.101193 (2023).
Kim, C. H. et al. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci. Lett. 216, 109–112. https://doi.org/10.1016/0304-3940(96)13021-4 (1996).
Bresciani, E., Broadbridge, E. & Liu, P. P. An efficient dissociation protocol for generation of single cell suspension from zebrafish embryos and larvae. MethodsX 5, 1287–1290. https://doi.org/10.1016/j.mex.2018.10.009 (2018).
Costa, K. H. A. et al. Ganglion cells and displaced amacrine cells density in the retina of the collared peccary (Pecari tajacu). PLoS One 15, e0239719. https://doi.org/10.1371/journal.pone.0239719 (2020).
Boff, J. M. et al. The interplay between neurotransmitters and calcium dynamics in retinal synapses during development, health, and disease. Int. J. Mol. Sci. 2024, 25. https://doi.org/10.3390/ijms25042226(2024).
Herrmann, R. et al. Rod vision is controlled by dopamine-dependent sensitization of rod bipolar cells by GABA. Neuron 72, 101–110. https://doi.org/10.1016/j.neuron.2011.07.030 (2011).
Agostinone, J. & Di Polo, A. Retinal ganglion cell dendrite pathology and synapse loss: Implications for glaucoma. Prog. Brain Res. 220, 199–216. https://doi.org/10.1016/bs.pbr.2015.04.012 (2015).
Oh-Hora, M. et al. Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat. Immunol. 9, 432–443. https://doi.org/10.1038/ni1574 (2008).
Berna-Erro, A. et al. STIM2 regulates capacitive Ca2+ entry in neurons and plays a key role in hypoxic neuronal cell death. Sci. Signal 2, ra67. https://doi.org/10.1126/scisignal.2000522(2009)
Boulton, M. & Dayhaw-Barker, P. The role of the retinal pigment epithelium: topographical variation and ageing changes. Eye (Lond.) 15, 384–389. https://doi.org/10.1038/eye.2001.141 (2001).
Heo, D. K., Lim, H. M., Nam, J. H., Lee, M. G. & Kim, J. Y. Regulation of phagocytosis and cytokine secretion by store-operated calcium entry in primary isolated murine microglia. Cell Signal 27, 177–186. https://doi.org/10.1016/j.cellsig.2014.11.003 (2015).
Berry, C. T., May, M. J. & Freedman, B. D. STIM- and Orai-mediated calcium entry controls NF-κB activity and function in lymphocytes. Cell Calcium 74, 131–143. https://doi.org/10.1016/j.ceca.2018.07.003 (2018).
Berna-Erro, A., Jardin, I., Salido, G. M. & Rosado, J. A. Role of STIM2 in cell function and physiopathology. J. Physiol. 595, 3111–3128. https://doi.org/10.1113/jp273889 (2017).
Moccia, F. et al. Stim and Orai proteins in neuronal Ca(2+) signaling and excitability. Front. Cell Neurosci. 9, 153. https://doi.org/10.3389/fncel.2015.00153 (2015).
Irnaten, M., O’Malley, G., Clark, A. F. & O’Brien, C. J. Transient receptor potential channels TRPC1/TRPC6 regulate lamina cribrosa cell extracellular matrix gene transcription and proliferation. Exp. Eye Res. 193, 107980. https://doi.org/10.1016/j.exer.2020.107980 (2020).
Arjun McKinney, A., Petrova, R. & Panagiotakos, G. Calcium and activity-dependent signaling in the developing cerebral cortex. Development 2022, 149. https://doi.org/10.1242/dev.198853 (2022).
Rosenberg, S. S. & Spitzer, N. C. Calcium signaling in neuronal development. Cold Spring Harb. Perspect. Biol. 3, a004259. https://doi.org/10.1101/cshperspect.a004259 (2011).
Morita, T., Matsumoto, S. & Baba, O. Expression of secretory calcium-binding phosphoprotein (scpp) genes in medaka during the formation and replacement of pharyngeal teeth. BMC Oral Health 23, 744. https://doi.org/10.1186/s12903-023-03498-7 (2023).
Rosa, J. T., Witten, P. E. & Huysseune, A. Cells at the edge: The dentin-bone interface in zebrafish teeth. Front. Physiol. 12, 723210. https://doi.org/10.3389/fphys.2021.723210 (2021).
Gu, Q. et al. Spatiotemporal characteristics of the pharyngeal teeth in interspecific distant hybrids of cyprinid fish: Phylogeny and expression of the initiation marker genes. Front. Genet. 13, 983444. https://doi.org/10.3389/fgene.2022.983444 (2022).
Kravchik, M. V. et al. Biomineralization of trabecular meshwork in open-angle glaucoma. Dokl. Biochem. Biophys. 495, 307–310. https://doi.org/10.1134/s1607672920060083 (2020).
Prasov, L. et al. DDX58(RIG-I)-related disease is associated with tissue-specific interferon pathway activation. J. Med. Genet. 59, 294–304. https://doi.org/10.1136/jmedgenet-2020-107447 (2022).
Soda, N., Sakai, N., Kato, H., Takami, M. & Fujita, T. Singleton-Merten syndrome-like skeletal abnormalities in mice with constitutively activated MDA5. J. Immunol. 203, 1356–1368. https://doi.org/10.4049/jimmunol.1900354 (2019).
Gutman, I., Melamed, S., Ashkenazi, I. & Blumenthal, M. Optic nerve compression by carotid arteries in low-tension glaucoma. Graefes Arch. Clin. Exp. Ophthalmol. 231, 711–717. https://doi.org/10.1007/bf00919286 (1993).
Mc, L. J. & Ray, B. S. Soft glaucoma and calcification of the internal carotid arteries. Arch. Ophthal. 38, 154–158. https://doi.org/10.1001/archopht.1947.00900010159002 (1947).
Agostinone, J. et al. Insulin signalling promotes dendrite and synapse regeneration and restores circuit function after axonal injury. Brain 141, 1963–1980. https://doi.org/10.1093/brain/awy142 (2018).
Al Hussein, A. l., Awamlh, S., Wareham, L. K., Risner, M. L. & Calkins, D. J. Insulin signaling as a therapeutic target in glaucomatous neurodegeneration. Int. J. Mol. Sci. 2021, 22. https://doi.org/10.3390/ijms22094672(2021).
Song, B. J., Aiello, L. P. & Pasquale, L. R. Presence and risk factors for glaucoma in patients with diabetes. Curr. Diab. Rep. 16, 124. https://doi.org/10.1007/s11892-016-0815-6 (2016).
Mrugacz, M., Bryl, A. & Zorena, K. Retinal vascular endothelial cell dysfunction and neuroretinal degeneration in diabetic patients. J. Clin. Med. 2021, 10. https://doi.org/10.3390/jcm10030458 (2021).
Tanaka, M., Sawada, M., Yoshida, S., Hanaoka, F. & Marunouchi, T. Insulin prevents apoptosis of external granular layer neurons in rat cerebellar slice cultures. Neurosci. Lett. 199, 37–40. https://doi.org/10.1016/0304-3940(95)12009-s (1995).
Faiq, M. A. et al. Ocular manifestations of central insulin resistance. Neural Regen. Res. 18, 1139–1146. https://doi.org/10.4103/1673-5374.355765 (2023).
Zhang, I. X., Ren, J., Vadrevu, S., Raghavan, M. & Satin, L. S. ER stress increases store-operated Ca(2+) entry (SOCE) and augments basal insulin secretion in pancreatic beta cells. J. Biol. Chem. 295, 5685–5700. https://doi.org/10.1074/jbc.RA120.012721 (2020).
Molnár, G. et al. GABAergic neurogliaform cells represent local sources of insulin in the cerebral cortex. J. Neurosci. 34, 1133–1137. https://doi.org/10.1523/jneurosci.4082-13.2014 (2014).
Chen, W., Cai, W., Hoover, B. & Kahn, C. R. Insulin action in the brain: Cell types, circuits, and diseases. Trends Neurosci. 45, 384–400. https://doi.org/10.1016/j.tins.2022.03.001 (2022).
Barone, E. et al. Biliverdin reductase-A mediates the beneficial effects of intranasal insulin in alzheimer disease. Mol. Neurobiol. 56, 2922–2943. https://doi.org/10.1007/s12035-018-1231-5 (2019).
Jarrett, S. G., Lewin, A. S. & Boulton, M. E. The importance of mitochondria in age-related and inherited eye disorders. Ophthalm. Res. 44, 179–190. https://doi.org/10.1159/000316480 (2010).
Krizaj, D. & Copenhagen, D. R. Calcium regulation in photoreceptors. Front. Biosci. 7, d2023–d2044. https://doi.org/10.2741/a896 (2002).
Percie-du-Sert, N. et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLoS Biol. 18, e3000410. https://doi.org/10.1371/journal.pbio.3000410(2020).
Ahrens, M. B., Orger, M. B., Robson, D. N., Li, J. M. & Keller, P. J. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods 10, 413–420. https://doi.org/10.1038/nmeth.2434 (2013).
Matthews, M., Trevarrow, B. & Matthews, J. A virtual tour of the Guide for zebrafish users. Lab. Anim. (N.Y.) 31, 34–40. https://doi.org/10.1038/5000140 (2002).
Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. https://doi.org/10.1093/bioinformatics/bts635 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Feng, J. et al. A genetically encoded fluorescent sensor for rapid and specific in vivo detection of norepinephrine. Neuron 102, 745-761.e8. https://doi.org/10.1016/j.neuron.2019.02.037 (2019).
Kaminow, B., Yunusov, D. & Dobin, A. STARsolo: accurate, fast and versatile mapping/quantification of single-cell and single-nucleus RNA-seq data. bioRxiv. https://doi.org/10.1101/2021.05.05.442755 (2021).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304. https://doi.org/10.1038/s41587-023-01767-y (2024).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296. https://doi.org/10.1038/s41592-019-0619-0 (2019).
Phipson, B. et al. propeller: Testing for differences in cell type proportions in single cell data. Bioinformatics 38, 4720–4726. https://doi.org/10.1093/bioinformatics/btac582 (2022).
Alexa, A. R. J. topGO: Enrichment Analysis for Gene Ontology. https://doi.org/10.18129/B9.bioc.topGO, R package version 2.54.0 (2023).
Lam, J. et al. A Universal approach to analyzing transmission electron microscopy with ImageJ. Cells 2021, 10. https://doi.org/10.3390/cells10092177(2021).
Stringer, C. & Pachitariu, M. Cellpose3: One-click image restoration for improved cellular segmentation. BioRxiv. https://doi.org/10.1101/2024.02.10.579780 (2024).
Acknowledgements
We thank Dr. Katarzyna Misztal from the Microscopy and Cytometry Facility at IIMCB in Warsaw for help with FACS cell separation, Prof. Bożena Kamińska for allowing us to use 10X Chromium system at the Nencki Institute of Experimental Biology, Dr. Natalia Ochocka and Dr. Kacper Adam Walentynowicz for help with cell encapsulation using 10X Chromium system, Dr. Justyna Zmorzyńska for providing the protocol for immunostaining on cryosections and shared knowledge of how to cryo-section zebrafish larvae eyes, personnel at the Zebrafish Core Facility at IIMCB (IN-MOL-CELL infrastructure) for maintenance of the zebrafish lines, Ms. Dominika Dubicka for administrative support, and Dr. Michael Arends for editing the manuscript. Sofiia Baranykova was supported by the Visegrad Fund as a Scholarship Program (grant no. 52310296) while conducting the experiments in this work. Figure 8 was originally created with BioRender.com by Sofiia Baranykova.
Funding
This research was funded by the National Science Centre (no. 2016/23/B/NZ3/03142) to J.K.
Author information
Authors and Affiliations
Contributions
Conceptualization, formal analysis, investigation: L.M., S.B., J.K., R.K.G. Methodology, software, data curation: S.B., R.K.G, A.K, S.S.M., I.W., C.L.W., L.M. Writing-original draft: L.M., J.K. Writing-review and editing: L.M., S.B., S.F., I.W., R.K.G, J.K. Visualization: S.B., M.M., A.Sz., T.W., L.M. Supervision: J.K. Project administration: J.K., L.M. Funding acquisition: J.K. All authors read and agreed to the published version of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Baranykova, S., Gupta, R.K., Kajdasz, A. et al. Loss of Stim2 in zebrafish induces glaucoma-like phenotype. Sci Rep 14, 24442 (2024). https://doi.org/10.1038/s41598-024-74909-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-74909-0
Keywords
This article is cited by
-
Zebrafish: A Research Paradigm for Studying Human Diseases
Proceedings of the Zoological Society (2025)