Abstract
FAR-RED IMPAIRED RESPONSE 1 (FAR1) is a class of transposase-derived transcription factors that play a very important role in the initiation of the photosensitive pigment A (phyA) signaling pathway. Despite their importance, the understanding of the function of FAR1 genes in quinoa is still limited, especially regarding how they affect the spike sprouting response. Quinoa has gained global attention in recent years for its health benefits and potential for sustainable agriculture. In our study, the CqFAR1 gene set in quinoa was characterized using HMMER (PF03101) and BLAST analyses, and 87 genes were identified. The 87 CqFAR1 genes were systematically classified into five groups that showed a high degree of conservation in gene structure and motif composition. Tissue expression profiles of the CqFAR1 gene indicated that the CqFAR1 gene plays a key role throughout the growth and development of quinoa, especially at mid (leaf) and end (spike) stages. By RT-qPCR analysis, we observed significant differences in the expression of the CqFAR1 gene at different developmental stages. Notably, the CqFAR1 gene showed significant expression enhancement at the early stage of quinoa spike sprouting. The results are useful for understanding the role of the CqFAR1 gene in quinoa growth and development and provide theoretical support for quinoa breeding.
Similar content being viewed by others
Introduction
FAR-RED IMPAIRED RESPONSE 1 (FAR1) is derived from a family of homologous proteins of transposases, originally identified in the model plant Arabidopsis thaliana as an important component of the photoreceptor pigment A (phyA)-mediated far-red-light signaling pathway, and belongs to a family of transcription factors that are found in most angiosperms1,2. The FAR1 protein contains three main functional structural domains: the N-terminal C2H2 zinc finger structural domain with DNA-binding activity; the central possible core transposase structural domain; and the C-terminal SWIM zinc finger structural domain, which has a transcriptional activation function3. In addition, the FAR1 gene family plays various important roles in plant physiology and development, including participation in light signaling responses4, various stress responses4, regulation of chlorophyll biosynthesis5, ABA signaling6, regulation of the biological clock and flowering time7, and other key processes. In terms of light signaling response, the FAR1/FHY3 dimer directly binds to the promoter. It activates the expression of two key regulators, FHY1 and FHL, which are key regulators of nuclear translocation, thus indirectly affecting the nuclear translocation process of PhyA8. During plant growth and development, dimers formed by FAR1 and FHY3 promote the expression of the ARC5 promoter by directly binding to it, thereby activating the synthesis of a dynamin protein essential for chloroplast division9. In addition, FHY3 and FAR1 can participate in the regulation of starch synthesis by promoting the transcriptional activation of ISA2, which in turn regulates starch synthesis induced by daytime light10. Particularly importantly, disruption of FHY3 and FAR1 expression significantly attenuated the inhibitory effects of ABA on seed germination, seedling greening, and root elongation processes6. In addition, they can bind to the ABI5 promoter and promote its transcriptional activity, which in turn participates in the ABA signaling process6. So far, the FAR1 gene family has been analyzed and reported in many plant species. For example, in Arabidopsis thaliana11, Arachis hypogaea12, Zea mays13, and Solanum tuberosum14, 12, 24, 16, and 20 FAR1 family members were found respectively. However, there is no research on the analysis of the FAR1 gene or the whole gene family in quinoa. Therefore, it is necessary to further study the function of CqFAR1 gene family members.
After a long domestication process, most crops have lost their inherent dormancy characteristics15. Under the continuous rainy or humid conditions before harvest, seeds are easy to germinate prematurely on the mother plant, which is called spike sprouting16. Spike sprouting usually severely reduces crop yield and quality17. According to statistics, the global grain loss caused by spike sprouting is more than 1 billion US dollars every year. In order to effectively reduce yield loss, it is very important to study the genetic basis of seed dormancy and spike sprouting.
Quinoa (Chenopodium quinoa Willd.), commonly known as "fake grain," is a dicotyledonous plant under Amaranthaceae, with the characteristics of tetraploid (2n = 4x = 36) C3 crops. It has a common history of more than 8000 years with the Andean indigenous culture and is honored as the "mother of ten thousand valleys" for its unparalleled nutritional and cultural value18,19,20,21. It is especially suitable for quinoa planted in alpine mountainous areas. Its maturity coincides with the rainy season, which easily causes spike sprouting, which will not only seriously affect the yield and quality of quinoa, but also reduce its commodity value and market competitiveness. However, research on the mechanism of seed dormancy and spike sprouting is relatively scarce, which largely limits the process of quinoa breeding. Therefore, it is very important to determine whether the FAR1 gene plays a role in seed dormancy and spike sprouting of quinoa.
In this study, we screened and identified the CqFAR1 gene family using the quinoa genome, analyzed its gene structure, phylogenetic relationships, GO functional annotation, and chromosomal localization, and performed expression pattern analysis based on the expression data of this family of genes in quinoa, as well as analyzed the promoter cis-acting elements. In addition to light-responsive elements, the quinoa FAR1 gene also contains abundant hormone-responsive elements and growth and developmental elements. We investigated the expression trends of the CqFAR1 gene at different times and initially discussed the expression patterns of these family members under spike sprouting, to provide theoretical references and bases for subsequent functional studies and applications of the CqFAR1 gene in quinoa.
Results and analysis
Identification and physicochemical properties of CqFAR1 gene family members
A total of 87 CqFAR1 genes were identified from the quinoa genome (Table 1), and the protein sequence length encoded by the genes ranged from 138 to 1,051 amino acids, with an average protein length of 526 amino acids. The molecular weight ranged from 16,702.1 kDa to 121,696.35 kDa, and the average molecular weight was 60,470.21 kDa. AUR62033390 had the largest protein length and molecular weight, while AUR62026577 had the smallest. The isoelectric point is between 4.71 and 9.62, and the average isoelectric point is 7.04. The instability coefficient of most proteins was > 40, indicating that they were unstable proteins. The fat index ranged from 55.68 to 85.23, and the average fat index was 70.81. The total average hydrophilicity is < -0.2. According to the Protscale function in ExPASY, it is known that all proteins are hydrophilic proteins. In addition, the subcellular localization predicted that most of CqFAR1 was located in the nucleus, cytoplasm, and chloroplast, while only AUR62008862 was located in the cytoskeleton, and AUR62017651 was located in the peroxisome.
Phylogenetic analysis of CqFAR1 genes
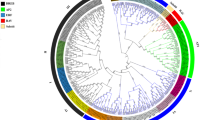
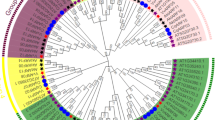
To deeply analyze the evolutionary relationship of the CqFAR1 gene family in quinoa, we constructed an unrooted phylogenetic tree based on the FAR1 protein sequences of C. quinoa, O. sativa, and A. thaliana. The CqFAR1 gene could be categorized into five main subgroups. The class I subfamily contains 18 genes, including 1 C. quinoa gene, 1 O. sativa gene, and 16 A. thaliana genes; there are 4 class II genes, including 0 C. quinoa genes, 2 O. sativa genes, and 2 A. thaliana genes; class III genes only contain 8 C. quinoa genes; there are 126 class IV genes, including 60 C. quinoa genes, 17 O. sativa genes, and 49 A. thaliana genes; there are 130 class V genes, including 18 C. quinoa genes, 44 O. sativa genes, and 68 A. thaliana genes (Fig. 1). These data show that the closer the phylogenetic relationship of family members, the higher the similarity of gene structure and domain. Further analysis suggested that different subtypes of FAR1 genes may assume different biological functions and genes with similar structures may play a related role in the regulation of growth and development of quinoa.
Conservative domain, gene structure and visual analysis of CqFAR1 protein
The composition and distribution of motifs are relatively conserved among members of the same subpopulation, and motifs between different subpopulations may lead to differences in CqFAR1 gene function (Fig. 2a). Motif 1 exists in all protein sequences. Motif 2 and Motif 3 are located at the N-terminal of most CqFAR1 protein sequences. The motif types and numbers of the same subgroup are similar, indicating that the motif pattern may be related to the function of the CqFAR1 protein. The conserved domain analysis of CqFAR1 protein using TBtools-II (v2.119) software showed that most genes belonged to the FAR1 superfamily and FHY3 superfamily. Only AUR62036755 had the DBD_Tnp_Mut superfamily, AUR62023657 had DUF4283, AUR62017462 had OTU_plant_OTU9-like, AUR62008862 had the FBD superfamily, and AUR62042833 had the MULE superfamily. These results showed that the structure of the CqFAR1 gene was diverse (Fig. 2b). To further understand the distribution of CqFAR1 exons and introns, clarify the inherent sequence diversity of the CqFAR1 gene family, and analyze its exon and intron distribution and cross-species conservation, the results showed that the number of exons in CqFAR1 genes ranged from 1 to 16. It is worth noting that AUR62024057 has only one exon, and AUR62017462 has 16 exons. The CDS of most CqFAR1 genes is not interrupted by introns. Although there are differences in the location of exons, the CqFAR1 gene family members clustered in the same phylogenetic branch usually have similar or the same number of exons (Fig. 2c).
Protein conserved motif, gene structure and visual analysis of quinoa CqFAR1 gene family. (a) The analysis of conserved motifs in FAR1 proteins. In total, three different motifs are represented by squares of different colors. (b) Analysis of protein domains in FAR1 proteins. Various symbols represent different protein structural domains. (c) Exon–intron structures of CqFAR1 genes. Exon–intron structures of CqFAR1 genes were obtained after analysis with TBtools-II (v2.119) for gene structure. Red bars indicate upstream and downstream UTRs, blue bars indicate coding sequences (CDS), and black lines indicate introns in the gene diagrams.
Chromosomal localization and gene duplication of CqFAR1 genes
Chromosomal localization results showed that the CqFAR1 gene family was distributed on all chromosomes except chromosome 13. Among them, the number of CqFAR1 genes on chromosome 17 and chromosome 1 was the most (12 and 9, respectively), while the number of CqFAR1 genes on chromosomes 8, 12, and 14 was the least, with only one CqFAR1 gene. In other chromosomes, there are 8 CqFAR1 genes on chr00, 4 genes on chr06, 8 genes on chr09, 2 genes on chr10, 4 genes on chr11, 3 genes on chr02, chr04 and chr15, 6 genes on chr03, chr07 and chr16, and 5 genes on chr05 and chr18. CqFAR1 genes on some chromosomes tend to exist in clusters, such as AUR62003731, AUR62003777, AUR62003863, AUR62003871 and AUR62003900, which are closely adjacent in physical position. On the whole, CqFAR1 genes tend to be located at the bottom of the chromosome, followed by the top, and the middle region is less populated. In addition, most CqFAR1 genes tend to be located in chromosome regions with high gene density (Fig. 3a). Gene duplication plays an important role in the evolution of organisms. The replicated genes lay the foundation for the physiological and morphological changes of individual plants. The whole genome of quinoa was analyzed for collinearity. The results are as follows (Fig. 3b). A total of 19 collinearity gene pairs were identified among 87 CqFAR1 genes. These collinearity gene pairs were scattered across 18 chromosomes of quinoa. There may be a large number of gene duplication events in the evolution of CqFAR1 family genes. In the quinoa genome, the CqFAR1 gene family has been amplified to some extent. All these indicate that gene duplication plays a very important role in the evolution of quinoa.
Collinearity analysis of CqFAR1 gene
Collinearity analysis is an important tool to reveal the process of genome differentiation and evolution. To further study the covariance of the CqFAR1 gene family, we found a collinear relationship between CqFAR1 in C. quinoa, O. sativa and A. thaliana (Fig. 4). One pair of CqFAR1 gene and OSFAR1 gene had collinearity (AUR62017462-OS04T0402300-00), and 10 pairs of CqFAR1 gene and AtFAR1 gene had collinearity (AUR62023864-AT2G43280.1, AT3G59470.2, AUR62011505-AT1G80010.1, AT1G52520.1, AUR62016322-AT1G80010.1, AT1G52520.1, AUR62013200-AT4G38170.1, AUR62002320-AT2G43280.1, AT3G59470.2, AUR62001611-AT2G27110.1), indicating that the quinoa CqFAR1 gene family is more closely related to the A. thaliana FAR1 gene family and may have similar biological functions.
Cis acting elements in the promoter region of CqFAR1 gene
Cis-acting elements are involved in regulating gene expression. We identified 65 cis-regulatory elements in the promoter region of the CqFAR1 gene family, which were mainly divided into plant hormone elements (abscisic acid response element, salicylic acid response element, gibberellin-response element, cis-acting regulatory element involved in the MeJA-responsiveness, auxin-responsive element); environmental stress elements (cis-acting regulatory element essential for the anaerobic induction, cis-acting element involved in low-temperature responsiveness, cis-acting element involved in defense and stress responsiveness, cis-acting regulatory element involved in circadian control, MYB binding site involved in drought-inducibility); photoresponse elements (light responsive element, MYB binding site involved in light responsiveness, part of gapA in (gapA-CMA1) involved with light responsiveness); growth and development elements (cis-regulatory element involved in endosperm expression, cis-acting regulatory element involved in seed-specific regulation, AT-rich DNA binding protein (ATBP-1), MYBHv1 binding site, cis-acting regulatory element related to meristem expression, involved in endosperm-specific negative expression, protein binding site, element involved in differentiation of the palisade mesophyll cells, cis-acting element involved in cell cycle regulation, cis-acting regulatory element root specific, wound-responsive element); secondary metabolic elements (cis-acting regulatory element involved in zein metabolism regulation, MYB binding site involved in flavonoid biosynthetic genes regulation) (Fig. 5). Most CqFAR1 genes exhibit at least one plant hormone response element in their promoter region. Ten cis-acting elements of phytohormone response were identified, including abre, auxrr core, TGA element, TGA box, p-box, gale motif, TatC box, tgacg motif, CGTCA motif, and TCA element. Cis-acting elements play an important role in regulating the reactivity of ABA, auxin, gibberellin, methyl jasmonate, and salicylic acid. At the same time, the abre homeopathic element is the most common in the CqFAR1 gene family. In addition, photoresponse elements were present in the promoter region of all CqFAR1 genes, and 36 cis-acting elements played a role in photoresponse. Six cis-regulatory elements related to abiotic stress response were identified, including LTR, ARE, MBS, TC-rich repeats, circadian, and GC-motif. The root-specific regulatory elements related to growth and development only exist in the gene AUR62039343. In addition, the MYB binding site was found in AUR62003863, AUR62023657, and AUR62044420, which is an indispensable part of the regulation of flavonoid biosynthesis gene elements. In conclusion, the CqFAR1 gene family plays an important role in regulating plant growth and development.
GO annotation and functional differentiation of the CqFAR1 gene family
To understand the function of quinoa CqFAR1, we performed GO functional annotation of 87 CqFAR1 genes, of which 47 CqFAR1 genes were annotated to biological process (BP), molecular function (MF), and cellular component (CC) categories in the GO database. Among the 47 quinoa CqFAR1 genes that were functionally annotated, 33 quinoa CqFAR1 genes were annotated for biological processes, 11 quinoa CqFAR1 genes for molecular functions, and 3 CqFAR1 genes for cellular components (Fig. 6a). The GO enrichment bubble plot (Fig. 6b) indicated that the main enrichment was in positive regulation of circadian rhythms, cellular response to far-red light, negative regulation of the developmental process, and other metabolic pathways. These indicate that the CqFAR1 gene family has functional differentiation and multiple functions in quinoa.
Expression pattern of CqFAR1 gene
Gene expression patterns provide important clues for understanding potential gene functions. Most CqFAR1 family members are expressed in all; however, their expression levels in different strains are quite different. Some family members showed preferential expression, and most genes were highly expressed in the control group. Specifically, the expression levels of AUR62003731 and AUR62003871 genes were the highest in the control, while the expression levels of AUR62003777 and AUR62003863 genes were the highest at 12 h (Fig. 7a). These findings suggest that the CqFAR1 gene family members have potential roles in growth and development as well as dormancy response. CqFAR1 genes showed significant expression levels in quinoa spikes, especially the genes AUR62003731 and AUR62003871. In contrast, the expression of these genes was relatively low in leaves and lowest in quinoa grains. This finding provides important clues for further investigation of the role of CqFAR1 genes in the growth and development of quinoa (Fig. 7b).
Real-Time Fluorescence Quantitative PCR Validation
To further investigate the effect of the FAR1 gene on quinoa spike sprouting and to verify the accuracy of the transcriptomic data, we selected six representative FAR1 genes and five transcriptomic genes in quinoa for real-time fluorescence quantitative analysis (Fig. 8). The results showed that the expression trend of these 11 genes detected by RT-qPCR closely matched the expression trend in the RNA-seq data, proving the reliability of gene expression in this study. The RT-qPCR assay and expression pattern analysis revealed that, except for AUR62003731 and AUR62003871, the expression of the remaining four CqFAR1 genes was very low in both resistant and sensitive lines, and it was speculated that they were not key regulatory genes. In addition, the decreasing trend of the expression of AUR62003731 and AUR62003871 in the sensitive lines was significantly higher than that in the resistant quinoa lines, suggesting that there is some kind of resistance mechanism in the resistant lines, so that the expression of AUR62003731 and AUR62003871 can be maintained or only slightly affected. In conclusion, it can be seen that the FAR1 gene is responsive to quinoa spike sprouting.
Discussion
Gene family analysis reveals the relationship between genetic diversity and functional evolution, providing us with a perspective to deeply understand the role of genes in the adaptive evolution of species22. In this study, we identified 87 CqFAR1 genes in quinoa, which is significantly more than other crops. This may be due to the combined effect of multiple factors such as quinoa genome size and complexity23, gene duplication and amplification24, genome doubling events25, and differences in bioinformatics analysis methods26. The quinoa FAR1 gene is distributed on 18 chromosomes, and 19 pairs of genes have tandem replication and segmental replication, indicating that genome replication events play an important role in maintaining the function of the gene family27. The reason for this analysis may be the high number of quinoa CqFAR1 gene family members. Based on the subcellular localization of CqFAR1 family proteins, most of the CqFAR1 proteins were localized in the cytoplasm. In O. sativa, the majority of OsFAR1 predictions showed that the gene was localized in the cytoplasm28. In A. thaliana, the majority of AtFAR1 predictions showed that the gene was localized in the cytoplasm and endoplasmic reticulum22. The prediction results in quinoa were largely consistent with those in O. sativa and A. thaliana, and it is hypothesized that the CqFAR1 gene may perform similar biological functions at the same site in different species. After analysis, it was found that there were some differences in gene structure and motif of different subgroups. They all contain the FAR1 domain. Phylogenetic tree analysis revealed that quinoa could be divided into five groups, and each group member showed similar exon length and number as well as motif structure. This finding showed that genes belonging to the same group were similar in function.
FAR1 family genes not only have structural diversity, but also may have other functional characteristics. These genes contain a variety of homeopathic elements29, which enable them to respond not only to light signals, but also to the adaptation of quinoa under hypoxia and low temperature conditions and the regulation of circadian rhythm. In addition, the abundant hormone response elements in this family mean that they may play a role in the hormone signal transduction pathway of quinoa. In the promoter region of quinoa CqFAR1 gene, several cis-regulatory elements have been identified, including GATA element, Box-4 and G-box. These findings suggest that CqFAR1 gene may be involved in the photosensitive reaction process in quinoa30. There is often a precise regulatory relationship between light reaction and a variety of hormones, which interact with each other to maintain physiological balance3. In this study, CqFAR1 gene, as a positive regulator of ABA signal transduction, has been further revealed. Studies have shown that the promoter of this gene contains multiple response elements, such as ABRE, AUXRR core, TGA element, TGA box and P-box, which may be closely related to ABA synthesis and signal transduction process, indicating that CqFAR1 plays a key role in the ABA signaling pathway31.
Induction of dormancy is a prerequisite for the initiation of dormancy in plants. PhyA, as a major photoreceptor of far-red light, induces the expression of a series of far-red light-responsive genes by transferring them from the cytoplasm to the nucleus, which in turn regulates processes such as seed germination and seedling de-yellowing. In the model plant A. thaliana, far-red light significantly down-regulated the expression levels of FAR1 and FHY38. As key regulators in the far-red light pathway, FAR1 and FHY3 are not only involved in light signaling but also interact with the abscisic acid (ABA)-related transcription factor ABI5, thus linking the light signaling pathway and the hormone signaling pathway, which together affect the dormant state of seeds6. It is noteworthy that the expression of FAR1 and FHY3 can also be induced by ABA, which further reveals the complex interaction mechanism between light signaling and hormone signaling6. In this study, we found that the expression of AUR62003731 and AUR62003871 decreased consistently throughout spike sprouting, which may be involved in the induction process of quinoa spike sprouting. In different strains of quinoa, the expression patterns of the CqFAR1 gene at the same time showed significant differences. In particular, the gene showed a high expression level in the spike of most strains, which suggested that the CqFAR1 gene might play a key role in the growth and development of the spike. By analyzing the expression patterns of genes, we can predict the physiological activities of these genes to some extent. Previous studies have revealed that in A. thaliana, members of the FAR1 gene family play a key role in regulating flowering timing and growth and development32. In this study, we found that the CqFAR1 gene had a significant expression pattern in spike tissues. However, the expression of this gene showed a gradual decrease as the spike sprouting process progressed. It is noteworthy that the expression level of CqFAR1 gene was significantly higher in the sensitive spike sprouting lines compared to the resistant spike sprouting lines. The regulation of the FAR1 gene on agronomic traits such as spike sprouting has been confirmed in other plants. In A. thaliana and O. sativa, the FAR1 gene regulates the synthesis and accumulation of phytochrome by interacting with the FHY1 protein. These genes play a crucial role in the light signal transduction pathway of plants, affecting the perception and adaptation of plants to light conditions33,34. This study showed that the expression level of the CqFAR1 gene in quinoa panicle was higher than that when the panicle had sprouted, indicating that CqFAR1 may be involved in regulating the process of panicle germination.
Conclusion
In this study, 87 members of the CqFAR1 gene family were identified. After examining their physicochemical properties, chromosomal localization, gene organization, evolutionary relationships, and covariance, it was determined that the 87 quinoa CqFAR1 gene family members were distributed on 18 chromosomes. The CqFAR1 genes were categorized into five subfamilies, with each subgroup exhibiting the same gene structure and motif. How the quinoa CqFAR1 gene family is expressed in various tissues and how it responds to spike sprouting was investigated. AUR62003731 and AUR62003871 showed significant differences in different quinoa lines and different tissue sites. The findings of this study provide important insights into the evolution of the CqFAR1 gene family and provide a solid foundation for further exploration of the specific functions of its family members.
Materials and methods
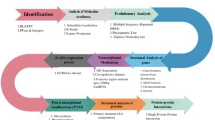
Data source and sequence acquisition of gene family analysis
From ensemblplants35 (http://plants.ensembl.org/index.html) download the genome files (cDNA, CDS, DNA.toplevel, PEP) and annotation files (GTF, GFF3) of C. quinoa and O. sativa. From TAIR (https://www.arabidopsis.org/), download the genome and annotation files of A. thaliana FAR1.
Gene family identification
To identify all FAR1 gene family members in C. quinoa, first, a TBtools-II (v2.119)36 Blast search (score ≥ 100, e ≤ 1e-10) was performed on the genome-wide protein files of C. quinoa based on the A. thaliana FAR1 protein sequence to obtain the candidate FAR1 genes. Second, the hidden Markov model file of the FAR1 domain (FAR1, PF03101) was downloaded from the Pfam database37 (http://pfam.xfam.org/), and the HMMER (v3.2.1) program in TBtools-II (v2.119)36 was used to search the whole genome protein of C. quinoa to obtain the candidate FAR1 gene. Third, SMART38 (http://smart.embl-heidelberg.de/) and batch CD search39 (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) were used to domain test all candidate FAR1 proteins searched by BLAST and HMMER, and each candidate gene was artificially screened to obtain a complete FAR1 protein. Finally, the online tool program on the ExPASy40 website (https://web.expasy.org/protparam/) was used to analyze the physicochemical properties of the FAR1 protein, including protein length, molecular weight, pI, instability index, aliphatic index, grand average hydrophilicity. The Wolf PSORT41 website (https://wolfpsort.hgc.jp/) was used to predict the subcellular localization of the FAR1 protein.
Phylogenetic evolutionary analysis of the FAR1 gene family in C. quinoa
The sequences of the identified FAR1 proteins were subjected to phylogenetic analysis with those of the A. thaliana FAR1 family and O. sativa FAR1 family protein sequences using the built-in Clustal W program in MEGA 7.0 software42. The phylogenetic tree was constructed using the neighbor-joining (NJ) method, with the Poisson model selected and the Bootstrap value set to 1,000, the missing values handled by pairwise deletion, and other parameters using the default values. To enhance the visualization, the phylogenetic tree was beautified using EvolView43 (https://www.evolgenius.info/evolview/).
Analysis of gene structure and conserved structural domains of the FAR1 gene family in C. quinoa
The MEME Suite online tool44 was used to analyze the conserved structural domains of the CqFAR1 gene family members with the parameter number of motifs set to 3, and the TBtools-II (v2.119)36 software was used to draw the gene structure and motif map. Motif analysis was performed on the protein conservative sequence of each FAR1 (motif number is 3, and the rest is the default). Finally, TBtools-II (v2.119)36 was used to organize and visualize the gene structure of FAR1 family members.
Chromosome localization and collinearity analysis of the FAR1 gene family in 5.6 C. quinoa
Firstly, this study needs to obtain the gene structure information and chromosomal localization data of the C. quinoa FAR1 transcription factor family, which can be extracted from the genome annotation files (GFF3, GTF). In TBtools-II (v2.119)36 software, the “Gene Density Profile” function was used to generate a gene density file, which provided the necessary data support for the mapping of gene localization. Then, the chromosome names and lengths were extracted in “Fasta Tools.” Finally, the “Gene Location Visualization” function was used to visualize the gene locations.
Chenopodium quinoa FAR1 gene family duplicates were identified based on the following criteria: (1) coverage of the shorter gene sequence length was greater than 70% of the longer sequence; and (2) similarity was greater than 70% when comparing the two sequences. Two genes of the same family that are five or fewer genes apart in a 100 kb chromosome segment are considered tandem duplications45,46. Next, to deeply understand the evolutionary relationship of FAR1 family genes, this study analyzed the collinearity of FAR1 family genes in C. quinoa, A. thaliana, and O. sativa using the MCScanX algorithm in TBtools-II (v2.119)36. Subsequently, the visualization tools in TBtools-II (v2.119)36 were used to intuitively display the analysis results to facilitate the comparison and interpretation of the collinearity evolution patterns of FAR1 family genes among different species.
Analysis of promoter cis-acting elements of the CqFAR1 gene family in C. quinoa
First, we extracted all upstream sequences (2000 bp) of the quinoa FAR1 gene family and uploaded them to the PlantCARE47 database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/). After manually screening for relevant cis-acting elements, we mapped them using the TBtools-II (v2.119)36 software.
GO annotation and functional classification of the FAR1 gene family in quinoa
Blast2Go version 5.048 was used to annotate and classify FAR1 transcripts. According to the annotation results, these transcripts were divided into different functional categories (biological process, molecular function, and cellular composition), and the number of transcripts under each functional category was counted. The results were visualized using the online tool jvenn (https://jvenn.toulouse.inrae.fr/app/example.html)49.
Analysis of plant materials, growth conditions, treatments, and tissue site expression patterns
It was planted in Xundian County, Kunming City, Yunnan Province (102°41′E, 25°20′N, 1766 m above sea level) and harvested immediately after 120 days of maturity using conventional management and normal fertilization. Since quinoa spikes had more seeds and were covered with sepals, which were difficult to observe, one spikelet was randomly selected from the main spike of quinoa and clipped for the experiment. Two quinoa lines, sensitive (Dianli-222) and resistant (Dianli-654), were selected. Distilled water was sprayed regularly to provide a moist environment for quinoa. When determining spike sprouting, the number of seeds was observed and counted every 6 h. The results showed that the sensitive strain partially germinated at 6 h, the resistant strain showed no sprouting, and the difference was greatest at 12 h, which was the optimal sampling time for both strains (0/6/12 h). A total of 18 samples with three biological replicates were frozen in liquid nitrogen and stored in a − 80 °C refrigerator for subsequent transcriptome sequencing analysis. The experimental process of transcriptome sequencing includes RNA extraction, RNA detection, library construction, and online sequencing. The downstream data will be filtered to obtain clean data, and then compared with the specified reference genome to obtain mapped data, which will be used to perform structural level analyses such as variable splicing analysis, new gene discovery, and gene structure optimization, etc. According to the expression of the genes in different samples or different sample groups, we will carry out expression level analyses such as differential expression analysis, differentially expressed gene function annotation, and function enrichment analysis. We used FPKM (Fragments Per Kilobase of transcript per Million fragments mapped) as a measure of the expression level of transcripts or genes, and screened differential genes with |log2Fold Change|≥ 1 and FDR < 0.05. We used RNA-seq data (https://www.ncbi.nlm.nih.gov/sra/SRP474959), including sensitive and resistant material, corresponding to three periods. We analyzed the transcript levels of the FAR1 gene family in different strains during the same period and used TBtools-II (v2.119)36 to create a heat map of gene expression (v2.119). In addition, leaf, seed, and spike transcriptome sequencing data of quinoa were downloaded from the NCBI SRA database (http://www.ncbi.nlm.nih.gov/sra/) under the sequence numbers SRP383884, SRP474959, and SRP431581, respectively. Based on the above data, the transcriptome sequencing data of C. quinoa were analyzed using Tophat50 and Cufflink51 software packages to analyze the transcriptome sequencing data and perform expression calculations to extract the gene expression data of the FAR1 gene family members. The expression data were normalized, and the expression heatmaps created using TBtools-II (v2.119)36 software illustrated the expression patterns of the C. quinoa FAR1 gene family in various tissues.
Presentation of RNA and establishment of cDNA libraries
In this experiment, the CTAB method was used to extract RNA52. The kit used was the Hieff NGS® Ultima Dual-mode mRNA Library Prep Kit. The library construction process included: total starting RNA ≥ 1 µg, enrichment of mRNA and fragmentation using Oligo(dT) magnetic beads; synthesis of the first strand of cDNA by M-MuLV reverse transcriptase, and synthesis of the second strand by RNase H degradation of RNA; after double-stranded cDNA was end-repaired, A-tailed ligated and ligated with sequencing junctions, the cDNA library was screened, PCR-amplified and purified. The cDNA library was initially quantified and diluted to 1.5 ng/μL using Qubit 2.0, the insert size was measured by Agilent 2100, and the effective concentration (> 2 nM) was accurately quantified by qRT-PCR to ensure the quality.
Real-time fluorescence quantitative PCR validation
To verify the reliability of the transcriptome sequencing results, the FAR1-related genes with high expression in the transcriptome were selected for RT-qPCR experiments. The ACT-1 gene was chosen as an internal reference gene and primers for the associated gene were designed in Beacon Designer 7.9 for RT-qPCR analysis53,54. The PerfectStart SYBR qPCR supermix (TransGen Biotech, Beijing, China) was used for RT-qPCR according to the manufacturer’s instructions. The reaction volume was 20 µL, including 2 × Perfectstart™ SYBR qPCR Supermix 10 µL, calibration solution 0.4 µL, nuclease-free water (RNase-free water) 6.8 µL, forward primer and reverse primer each (10 µM) 0.4 µL, cDNA 2 µL (200 ng/µL). The thermal cycle was as follows: 94 °C (30 s), 94 °C (5 s), 60 °C (30 s), for 40 cycles. The relative gene expression level was calculated using the 2−ΔΔCT method55.
Data availability
The original contributions presented in the study are publicly available. This data can be found here National Center for Biotechnology Information (NCBI) SRA database under accession number a SRP474959. The names of the repository and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra/SRP474959.
References
Wang, H. & Wang, H. Multifaceted roles of FHY3 and FAR1 in light signaling and beyond. Trends Plant Sci. 20, 453–461 (2015).
Jiang, Y. et al. Genome-wide identification and integrated analysis of the FAR1/FHY3 gene family and genes expression analysis under methyl jasmonate treatment in Panax Ginseng C A. Mey. BMC Plant Biol. 24, 549 (2024).
Xie, Y. et al. Arabidopsis FHY3 and FAR1 integrate light and strigolactone signaling to regulate branching. Nat. Commun. 11, 1955 (2020).
Wang, H. & Deng, X. W. Arabidopsis FHY3 defines a key phytochrome a signaling component directly interacting with its homologous partner FAR1. Embo J. 21, 1339–1349 (2002).
McCormac, A. C. & Terry, M. J. Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in arabidopsis Thaliana. Plant J. 32, 549–559 (2002).
Tang, W. et al. FAR-RED elongated HYPOCOTYL3 and FAR-RED IMPAIRED RESPONSE1 transcription factors integrate light and abscisic acid signaling in arabidopsis. Plant Physiol. 163, 857–866 (2013).
Li, G. et al. Coordinated transcriptional regulation underlying the circadian clock in arabidopsis. Nat. Cell Biol. 13, 616–622 (2011).
Lin, R. et al. Transposase-derived transcription factors regulate light signaling in arabidopsis. Science. 318, 1302–1305 (2007).
Ouyang, X. et al. Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in arabidopsis development. Plant Cell. 23, 2514–2535 (2011).
Ma, L., Xue, N., Fu, X., Zhang, H. & Li, G. Arabidopsis Thaliana FAR-RED ELONGATED HYPOCOTYLS3 (FHY3) and FAR-RED-IMPAIRED RESPONSE1 (FAR1) modulate starch synthesis in response to light and sugar. New Phytol. 213, 1682–1696 (2017).
Dai, J. et al. FAR1/FHY3 transcription factors positively regulate the salt and temperature stress responses in eucalyptus grandis. Front. Plant Sci. 13, 883654 (2022).
Lu, Q. et al. Genome-wide identification and expression of FAR1 gene family provide insight into pod development in peanut (Arachis hypogaea). Front. Plant Sci. 13, 893278 (2022).
Tang, H. et al. Genome-wide identification and expression analyses of the FAR1/FHY3 gene family provide insight into inflorescence development in maize. Curr. Issues Mol. Biol. 46, 430–449 (2024).
Chen, Q. et al. Genome-wide identification and functional characterization of FAR1-RELATED SEQUENCE (FRS) family members in potato (Solanum tuberosum). Plants (Basel). 12, (2023).
Liang, Y., Liu, H. J., Yan, J. & Tian, F. Natural variation in crops: Realized understanding, continuing promise. Annu. Rev. Plant Biol. 72, 357–385 (2021).
Park, M., Shin, S. Y., Moon, H., Choi, W. & Shin, C. Analysis of the global transcriptome and miRNAome associated with seed dormancy during seed maturation in rice (Oryza Sativa L. Cv. Nipponbare). BMC Plant Biol. 24, 215 (2024).
Lee, C. M. et al. QTL mapping and improvement of pre-harvest sprouting resistance using japonica weedy rice. Front. Plant Sci. 14, 1194058 (2023).
Ozcaliskan, I. H. et al. Quinoa (Chenopodium Quinoa Willd) supplemented cafeteria diet ameliorates glucose intolerance in rats. Food Sci Nutr. 11, 6920–6930 (2023).
Ain, Q. T. et al. Adaptive mechanisms in quinoa for coping in stressful environments: An update. PeerJ. 11, e14832 (2023).
Shen, Z. J. et al. TMT proteomics analysis of a pseudocereal crop, quinoa (Chenopodium Quinoa Willd) during seed maturation. Front. Plant Sci. 13, 975073 (2022).
Matias, J. et al. Assessment of the changes in seed yield and nutritional quality of quinoa grown under rainfed mediterranean environments. Front. Plant Sci. 14, 1268014 (2023).
Lin, R. & Wang, H. Arabidopsis FHY3/FAR1 gene family and distinct roles of its members in light control of arabidopsis development. Plant Physiol. 136, 4010–4022 (2004).
Bennett, M. D. & Leitch, I. J. Nuclear DNA amounts in angiosperms: Targets trends and tomorrow. Ann Bot. 107, 467–590 (2011).
Van de Peer, Y., Maere, S. & Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10, 725–732 (2009).
Wendel, J. F., Jackson, S. A., Meyers, B. C. & Wing, R. A. Evolution of plant genome architecture. Genome Biol. 17, 37 (2016).
Li, L., Stoeckert, C. J. & Roos, D. S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003).
Kong, H. et al. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 50, 873–885 (2007).
Zhou, D. et al. Characterization of the fatty Acyl-CoA reductase (FAR) gene family and its response to abiotic stress in rice (Oryza sativa L.). Plants (Basel). 13, (2024).
Marand, A. P., Eveland, A. L., Kaufmann, K. & Springer, N. M. Cis-regulatory elements in plant development, adaptation, and evolution. Annu. Rev. Plant Biol. 74, 111–137 (2023).
Baek, K. et al. Introducing dunaliella LIP promoter containing light-inducible motifs improves transgenic expression in chlamydomonas reinhardtii. Biotechnol. J. 11, 384–392 (2016).
Wang, X. et al. ABRE-BINDING FACTORS play a role in the feedback regulation of aba signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol. 221, 341–355 (2019).
Ritter, A. et al. The transcriptional repressor complex FRS7-FRS12 regulates flowering time and growth in arabidopsis. Nat. Commun. 8, 15235 (2017).
Feng, Z. J. et al. Foxtail millet NF-Y families: Genome-wide survey and evolution analyses identified two functional genes important in abiotic stresses. Front. Plant Sci. 6, 1142 (2015).
Wang, W. et al. A pair of light signaling factors FHY3 and FAR1 regulates plant immunity by modulating chlorophyll biosynthesis. J. Integr. Plant Biol. 58, 91–103 (2016).
Cunningham, F. et al. Ensembl 2022. Nucleic Acids Res. 50, D988–D995 (2022).
Chen, C. et al. TBtools-II: A “One for All, All for One” bioinformatics platform for biological big-data mining. Mol. Plant. 16, 1733–1742 (2023).
Blum, M. et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354 (2021).
Letunic, I. & Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496 (2018).
Lu, S. et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268 (2020).
Wilkins, M. R. et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 112, 531–552 (1999).
Horton, P. et al. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 35, W585–W587 (2007).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027 (2021).
Subramanian, B., Gao, S., Lercher, M. J., Hu, S. & Chen, W. H. Evolview V3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275 (2019).
Bailey, T. L. et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Gu, Z., Cavalcanti, A., Chen, F. C., Bouman, P. & Li, W. H. Extent of gene duplication in the genomes of drosophila, nematode, and yeast. Mol. Biol. Evol. 19, 256–262 (2002).
Yang, S., Zhang, X., Yue, J. X., Tian, D. & Chen, J. Q. Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genomics. 280, 187–198 (2008).
Lescot, M. et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327 (2002).
Conesa, A. et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 21, 3674–3676 (2005).
Bardou, P., Mariette, J., Escudie, F., Djemiel, C. & Klopp, C. Jvenn: An interactive Venn diagram viewer. BMC Bioinformatics. 15, 293 (2014).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 25, 1105–1111 (2009).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Mainkar, P. et al. Development of Modified CTAB and trizol protocols to isolate high molecular weight (HMW) RNA from polyphenol and polysaccharides rich pigeonpea (Cajanuscajan (L.) Millsp. PLoS One. 18, 291949 (2023).
Tian, C. et al. Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. PLoS One. 10, e117569 (2015).
Chen, C. et al. Identification of reliable reference genes for quantitative real-time PCR Normalization in Pitaya. Plant Methods. 15, 70 (2019).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25, 402–408 (2001).
Acknowledgements
We wish to acknowledge the Wuhan Metware Biotechnology Co., Ltd., for Professional technical services. We also thank professor Peng Qin conducted with the experiments for their valuable help and guidance.
Funding
We gratefully acknowledge the financial support of the Academician Expert Workstation (202405AF140012) and the Yunnan Expert Workstation (202205AF150001) and the “Xingdian Talent” Industry Innovation Talent Program in Yunnan Province (XDYCCYCX-2022–0029).
Author information
Authors and Affiliations
Contributions
LBH wrote the original draft and performed the methodology. LYZ wrote the original draft and carried out the formal analysis. PZ did the conceptualization, and wrote, reviewed, and edited the manuscript. JNL carried out the formal analysis, performed the methodology, and visualized the data. LL collected the field samples and prepared the plant materials. HXL carried out the formal analysis and investigated the data. XQW and YTB and GFJ carried out the formal analysis and investigated the data. PQ supervised the data and carried out the project administration and funding acquisition. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, L., Zhang, L., Zhang, P. et al. Molecular characteristics and expression pattern of the FAR1 gene during spike sprouting in quinoa. Sci Rep 14, 28485 (2024). https://doi.org/10.1038/s41598-024-79474-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-79474-0