Abstract
Radix Astragali (RA) has been recognized for its therapeutic potential in allergic rhinitis (AR), yet its potential pharmacological mechanisms remain elusive. This study systematically investigated the physicochemical properties and biological activities of RA’s phytochemicals, aiming to elucidate their targets and mechanisms in AR treatment. We identified 775 potential targets of RA’s key phytochemicals and intersected these with 29,544 AR-related disease targets, pinpointing 747 shared therapeutic targets. A protein-protein interaction network analysis categorized these targets into five subclusters, with TNF, NFKB1, IKBKB, NFKBIA, and CHUK emerging as central nodes. Enrichment analysis revealed their roles in inflammatory and immune responses, particularly through the NF-κB, TNF, IL-17, Toll-like receptor, and NOD-like receptor signaling pathways. Molecular docking and dynamics simulations confirmed the strong binding affinity and stability of RA’s phytochemicals to these targets. In vivo, RA intervention effectively reversed the expression of key inflammatory markers in an IL-13-induced nasal mucosa inflammation model. Our findings suggest that RA’s multitargeted approach involves the modulation of critical inflammatory pathways, highlighting its therapeutic potential.
Similar content being viewed by others
Introduction
Allergic rhinitis (AR) is a chronic airway inflammatory disorder characterized mainly by nasal symptoms, including itchy nose, sneezing, rhinorrhoea and nasal congestion, which is caused by immunoglobulin E (IgE)-mediated inflammation of the nasal membranes after allergen exposure1. AR is a global health issue that affects 10–20% of the population worldwide2 and severely impairs patients’ lives, including their social activities, ability to learn in school, and work productivity3. Additionally, AR is linked to various inflammatory illnesses of the mucosa, including asthma, allergic conjunctivitis, and rhinosinusitis1. Although H1-antihistamines, antileukotriene medications, and intranasal glucocorticosteroids are used to alleviate the allergic symptoms1,4, patients are still unsatisfied for relapse after drug withdrawal5 and were compelled to seek complementary and alternative therapies.
Radix Astragali (RA), known as Huangqi, is a natural product with a history of medicinal and nutritional use dating back to Shen Nong’s Classic of Materia Medica in 200 AD. In traditional Chinese medicine, it is the most representative invigorating Qi medicine, which is widely used for anti-fatigue, delaying aging, and treating deficiency diseases6,7,8. Modern pharmacological studies have revealed RA’s immunomodulatory, anti-inflammatory, and antiviral activities6,9. Specifically, RA’s active ingredients, such as formononetin and kaempferol, have demonstrated anti-allergic effects by modulating inflammatory pathways and cytokine production in AR10,11. Despite RA’s recognized therapeutic potential, a comprehensive understanding of its molecular mechanisms in AR treatment remains elusive.
Network pharmacology represents a cutting-edge, integrative approach that enables a comprehensive and systematic dissection of the biological targets, molecular functions, and underlying mechanisms of bioactive compounds. In this study, we have uncovered the therapeutic potential of RA in modulating the expression of pivotal inflammatory biomarkers in a model of IL-13-induced nasal mucosa inflammation. Our investigation was conducted through a multi-faceted strategy that combined network pharmacology predictions with rigorous experimental validation to delineate the therapeutic mechanisms. Furthermore, we conducted a detailed analysis of the interactions between key compounds in RA and the core molecular players implicated in allergic rhinitis progression, utilizing molecular docking and molecular dynamics simulations to elucidate these interactions.
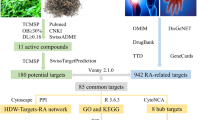
The graphical abstract encapsulating the essence of our findings is presented in Fig. 1.
Materials and methods
Physicochemical properties and biological activities of ingredients
The phytochemicals of RA were procured from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP), and oral bioavailability (OB) of ≥ 30% and a drug-likeness (DL) of ≥ 0.18 was set to candidate active ingredients. The Molinspiration server was utilized to assess the molecular descriptors, drug-likeness and bioactivity of the candidate active compounds. The value of the additional percentage of absorption (%ABS) was calculated as follows: %ABS = 109 – [0.345*TPSA]12.
Targets of RA and AR
TCMSP, Swiss Target Prediction, HERB, ChEMBL, PubChem, Search Tool for Interacting Chemicals (STITCH) and Super-PRED were used to find out targets of RA. The UniProt database was utilized to transform candidate targets into standard gene symbols with human settings, and then duplicates were removed. AR-associated targets were gathered from the eight open-source databases, including GeneCards, Online Mendelian Inheritance in Man (OMIM), Comparative Toxicogenomics Database (CTD), DisGeNET, Drugbank, MalaCards, NCBI Gene and Therapeutic Target Database (TTD). After eliminating duplications, all the disease targets were discovered. The ultimate targets of RA against AR were filtered by intersecting AR and RA targets.
Construction of PPI network
The STRING database was utilized to construct the protein-protein interaction (PPI) network of the shared targets with the setting was restricted to “Homo sapiens” and interaction scores greater than 0.99. Subsequently, Cytoscape was utilized to visualize and analyze the PPI network. The significant module of the network was screened by the MCODE plugin13.
Analysis of GO and KEGG pathway enrichment
Enrichment analysis of Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway14,15,16 were performed by in R with “ClusterProfiler” and “org.Hs.eg.DB”. Considering GO enrichment analysis may include a substantial amount of redundant information, ‘rrvgo’ was utilized to simplify the enrichment results17.
Molecular docking and molecular dynamics simulation verification
The 3D structure of the main phytochemical was obtained from the PubChem database. The core target’s crystal structure was downloaded from the Protein Data Bank database (PDB). PyMOL (2.4.0) was used to extract the crystal structure of the complex’s ligands and water molecules, and add hydrogen. Autodock tools were used to convert the pdb format into the pdbqt format, and AutoDock Vina performed the molecular docking between the receptor and ligand, with the final docking results visualized using PyMOL. The Schordinger 2019 software “Desmond” module was used for molecular dynamics (MD) simulation. A predefined Simple Point Charge (SPC) water model was used to simulate water molecules using the OPLS 2005 force field. In order to neutralize the system charge, an appropriate amount of chloride or sodium ions were added to balance the system charge and randomly placed in the solventized system. Performed energy minimization of the solvated system using the default protocol integrated with the Desmond module (using OPLS 2005 force field parameters). Employed Nose-Hoover temperature coupling and isotropic scaling to maintain the temperature and pressure at 300 K and 1 atmosphere; following the operations, ran a 100 ns NPT simulation, saving trajectories at 100 ps intervals.
Human nasal epithelial cells culture
Human nasal epithelial cells (HNEpCs) were purchased from Guangzhou Genio Biotech Co., Ltd. and have been confirmed to be consistent through STR identification. HNEpCs were cultured in DMEM supplemented with 18% fetal bovine serum and 100 U/mL penicillin, 100 μg/mL streptomycin in a 37 ℃ with 5% CO2 humidified incubator. When the cell growth reached confluence of 80% ~ 90%, they were digested at a ratio of 1:2 using 0.25% EDTA trypsin at 37 ℃. These cells were then seeded into a six-well plate until they reached a density of 90%. HNEpCs were incubated with IL-13 (100 ng/mL) for 12–24 h separately.
Preparation of Radix astragali lyophilized powder
The RA herbal medicine, which purchased from Kangmei Pharmaceutical Co., Ltd., was combined and pulverized using a high-speed universal machine. A 5-fold volume of deionized water was added to the mixture in a bottle. RA was soaked in deionized water for 30 min before being boiled twice at 100 °C in ten times its volume of deionized water for 2 h. The resulting filtrates were concentrated using a rotary evaporator and dried through lyophilization to obtain powdered extract, which was stored at 4 °C. Subsequently, the decoction process involved heating with a water frying sleeve thermostat for 120 min, followed by filtration of the drug residue through a 400 mesh filter screen and centrifugation at 3500 r/min (r = 40 cm) for 5 min. The supernatant containing the medicinal solution was then concentrated using a rotary evaporator until reaching a final concentration of 1 g/mL. Finally, the concentrate underwent freezing at -80℃ for 48 h before being freeze-dried with a vacuum freeze-drying machine over a period of 72 h.
CCK8
The logarithmically cultured HNEpCs were seeded into 96-well cell culture plates at a density of 1 × 104 cells per well. Various concentrations of freeze-dried Radix astragali powder (0 mg/mL, 0.16 mg/mL, 0.3125 mg/mL, 0.625 mg/mL, 1.25 mg/mL, 2.5 mg/mL, 5 mg/mL, 10 mg/mL and 20 mg/mL) were used to treat the cells for durations of both 24 h and 48 h. Subsequently, each well was supplemented with CCK8 reagent (10 μL) and incubated in darkness for a period ranging from half an hour to two hours. The absorbance at a wavelength of 490 nm was measured for each well.
Western blot
The proteins from each group were extracted using RIPA lysis buffer containing 1 mM phosphatase inhibitors and 1 mM protease inhibitors. After centrifugation of the cell lysates at 12,000 g and 4℃ for 15 min, the total protein content in the supernatant was quantified using the bicinchoninic acid protein assay kit. Supernatants were mixed with a 5×protein sample loading buffer, boiled for 10 min, and stored at -20℃ for future use. Subsequently, equal quantities of protein (40 μg/lane) from each sample were loaded onto a 10% SDS-PAGE gel and transferred to a polyvinylidene difluoride membrane with a pore size of 0.45 μm. The gel blocking step was performed using Tris-buffered saline containing 0.1% Tween-20 (TBST) and nonfat dry milk (5%) at room temperature for two hours, followed by overnight incubation with primary antibodies β-Actin/STAT3/ pSTAT3/ NF-κB / pNF-κB (diluted to 1:1000), which were purchased from CST, at a temperature of 4 ℃. After three washes with a TBST solution (10 min each), the membranes were incubated with HRP-conjugated goat anti-rabbit secondary antibody (diluted to 1:3000) for one hour at room temperature. Subsequent to three additional washes with TBST, chemiluminescence detection reagents were used to visualize the bands via the ChemiDoc Touch-1 (BIO-RAD).
Reverse transcription‑quantitative polymerase chain reaction (RT‑qPCR)
The HNEpCs were subjected to total RNA extraction using the Cell RNA Rapid Extraction Kit following the manufacturer’s guidelines. The extracted RNA was quantified using a Nano-300 Micro-Spectrophotometer, and samples with a D(λ)260/D(λ)280 ratio falling within 1.8–2.0 were stored at -80 °C. For cDNA synthesis, 1 μg of RNA was utilized with the HiScript RT SuperMix for qPCR (+ gDNA wiper) Kit. Reverse transcription was carried out at 50℃ for 15 min, followed by 85℃ for 5 s. All PCR primers were designed using NCBI Primer-BLAST and obtained from Guangzhou Miaobo Biotechnology (STable 1). RT-qPCR analysis was conducted on an ABI ViiA 7-384 instrument using ChamQ Universal SYBR qPCR master mix as per the following cycling conditions: initial denaturation at 95 ℃ for 30 s, followed by amplification cycles of denaturation at 95 ℃ for 5 s and annealing at 60 ℃ for 30 s. The expression levels of the GAPDH gene served as internal controls and were assayed simultaneously with the samples. Relative gene expression was determined utilizing the ΔΔCT method (2^−ΔΔCT). Each experiment was conducted in triplicate. Primer specificity validation included both in silico analysis (NCBI Primer-BLAST) and melting curve analysis post-qPCR amplification.
Results
Physicochemical properties and bioactivity of the active components
A comprehensive list of 87 phytochemicals was discovered in RA, with 20 active compounds meeting the inclusion criteria of oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18 (STable 2). We analyzed the physicochemical properties of 18 active ingredients in our study, excluding two that the PubChem database could not retrieve. Most key active phytochemicals, except MOL000433, MOL000374 and MOL000439, satisfied Lipinski’s Rule of Five with no violations. And the majority of active ingredients were in accordance with TPSA values < 140 Å2 and %ABS > 63%, indicating that they were within the optimal range of oral bioavailability (Table 1).
The physiological function for the key components of RA might be linked to various mechanisms, including the interplay of ion channel modulators, GPCR ligands, nuclear receptor ligands, kinase inhibitors, protease inhibitors and enzyme inhibitors. Molecular compounds with a bioactivity value greater than 0.00 have a high probability of exhibiting significant biological activity18. Mairin exhibited excellent enzyme inhibitor and nuclear receptor ligand affinities with bioactivity scores > 0.5, good protease inhibitor, ion channel modulator and GPCR ligand affinities with bioactivity values > 0, and moderate kinase inhibitor affinities with bioactivity scores ranging from − 5.0 to 0. The results illustrated that mairin possesses exceptional enzyme inhibitor and nuclear receptor ligand affinity (nuclear receptor ligand > enzyme inhibitor > GPCR ligand > protease inhibitor > ion channel modulator > kinase inhibitor). Table 2 presents comprehensive information on additional phytochemicals.
Identifying the targets associated with RA and AR
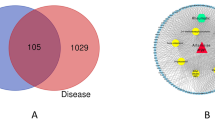
The targets of RA were extracted from seven open-source databases: TCMSP (209), CheMBL (255), HERB (233), PubChem (363), Swiss Target Prediction (375), STITCH (44) and Super-PRED (327). The RA-associated target set was acquired by merging the results of different databases, and 775 potential targets were acquired after eliminating duplicates (Fig. 2A). AR-associated targets were obtained via eight open-source databases: CTD (43837), GeneCards (2718), DisGeNET (446), NCBI gene (817), OMIM (3), DrugBank (18), TTD (30) and MalaCards (39). A total of 29,544 targets were obtained after the elimination of duplicates (Fig. 2B). Ultimately, a total of 747 intersection target was acquired as the target of RA against AR (Fig. 2C).
PPI network construction and top cluster analysis
The PPI was generated by using the STRING database with the 747 intersection targets (Fig. 2D, E). Figure 2F shows the top 30 ranked targets. MCODE was used to analyze the PPI network and recognized five significant clusters (Fig. 2G; Table 3). The top-ranked hub cluster including five key targets, namely TNF, NFKB1, IKBKB, NFKBIA and CHUK. Enrichment analysis showed that top-ranked hub cluster were involved in NF-kappa B signaling pathway, Toll-like receptor signaling pathway, T-cell receptor signaling pathway, TNF signaling pathway, IL-17 signaling pathway and so on (Fig. 2H, I).
(A) The number of target genes related to the ingredients of RA comes from seven open-source databases. (B) The number of target genes related to AR from eight open-source databases. (C) Venn diagram depicting common target genes between AR and RA. Results of PPI network analysis of Radix astragali interfering with AR intersection targets. (D) The PPI network of intersection targets. (E) PPI network visualized with Cytoscape; the color of each node represents the degree of freedom. (F) Bar chart of intersection targets with top 30 degree values. (G) Screened hub targets in Cytoscape with MCODE. (H) GO analysis of the top-ranked cluster. (I) KEGG analysis of the top-ranked cluster.
GO and KEGG pathway enrichment analyses of key clusters
For a more comprehensive understanding of the potential mechanisms of intervention, clusters with a score of four or higher were considered as important therapeutic targets, including clusters 1, 2, and 3 (Table 3). GO and KEGG analyses were also performed for these three clusters (Fig. 3A, B). Since the GO enrichment analysis might contain extensive duplicated data, the results were streamlined using an R package, as shown in Fig. 3C. The results showed that the GO biological processes mainly involved humoral immune response, regulation of epithelial cell proliferation, cellular response to lipopolysaccharide, cellular response to biotic stimuli, regulation of inflammatory responses, positive regulation of cytokine production, and cellular response to tumor necrosis factor, among others. The KEGG analysis revealed that the key targets were associated with 89 pathways, with the top 30 pathways displayed in a histogram and a bubble chart based on their counts or fold enrichment (Fig. 3D, E). The main KEGG pathways targeted by RA against AR included the NF-kappa B signaling pathway, TNF signaling pathway, IL-17 signaling pathway, Toll-like receptor signaling pathway, NOD-like receptor signaling pathway, and cytokine-cytokine receptor interaction, among others. The biological processes and KEGG pathways were strongly correlated with inflammation, immunological responses, and epithelial cell proliferation. Finally, we constructed an interaction network that includes the active ingredients of RA, targets, and KEGG pathways to better elucidate the multi-component and multi-target interventional role of RA in AR (Fig. 3F).
GO and KEGG enrichment analyses for main targets. (A) The bar plot shows the hub gene-enriched top 20 items ranked by counts. (B) The bubble diagram shows the hub genes-enriched top 20 items ranked by fold enrichment. (C) The rectangular tree diagram shows the simplified results of all the terms of GO enrichment. (D) The bar plot shows the hub gene-enriched top 20 KEGG enrichment pathways ranked by counts. (E) The bubble diagram shows the hub genes enriched in the top 20 KEGG enrichment pathways ranked by fold enrichment. (F) Network visualization from bioinformatics analysis highlighting the detailed interactions of ingredient-target-KEGG.
Binding activities of the key phytochemicals to core targets
The binding energy between the top-ranked clusters and their corresponding RA components was determined by molecular docking, as shown in Table 4. A lower binding energy implies a greater affinity. The docked complexes exhibited the binding affinities and interactions between ligand and receptor, as depicted in Fig. 4. Binding energies below − 5.0 kcal/mol are considered good binding interactions between the major compounds and targets19. The docking results indicated that the essential compounds of RA might bind to TNF, NFKB1, NFKBIA, IKBKB and CHUK. Furthermore, TNF (PDB ID, 2az5), NFKB1(7lfc) and NFKBIA (PDB ID, 6y1j) showed the lowest binding energies with MOL000387, MOL000378 and MOL000392, respectively.
Molecular dynamics simulations
Molecular docking revealed that MOL000417 (calycosin) displayed a superior binding energy to TNF compared to other compounds and proteins. Therefore, 100 ns molecular dynamics (MD) simulations were performed to investigate the stability of the calycosin-TNF complex. The stability of the protein-ligand complex was assessed using RMSD plots, which measured the deviation of protein and ligand atoms in the binding pocket at the end of the 100 ns simulation cycle relative to their initial positions at 0 ns before the simulation. In Fig. 5A, the RMSD plot of the complex’s 100 ns trajectory analysis is shown. The left Y-axis represents the RMSD change of TNF, while the right Y-axis represents the stability of calycosin relative to the protein and its binding pocket. It was observed that the calycosin-TNF complex exhibited some stability, with an RMSD of 1.4 Å for TNF and 9 Å for the complex. The complex demonstrated a highly fluctuating pattern before 55 s, after which it stabilized. The RMSF plot of TNF indicated that the residue with the highest flexibility and fluctuation during the simulation was at index 11, which is important for the protein’s functional performance (Fig. 5B). Interactions between calycosin and TNF residues were analyzed, revealing that interactions with TYR151, TYR119, TYR59, LEU57, and SER60 were maintained above 50%, 45%, 30%, 20%, and 25% of the simulated time, respectively (Fig. 5C). A detailed examination of interactions with protein residues indicated hydrophobic interactions with TYR151 (23%) and polar interactions with SER60 (14%) (Fig. 5D). The upper part of Fig. 5E illustrates the overall specific interaction between calycosin and TNF, while the lower part shows the protein residues that interact with calycosin at each time point. The dark orange color observed throughout the trajectory represents the interaction sites, including TYR151, TYR119, TYR59, LEU57, and SER60. It was found that these interactions primarily involved hydrogen bonding, hydrophobicity, and water bridges.
Dynamics were simulated in TNF and MOL000417 (Calycosin). (A) RMSD plot for the trajectory analysis of the complex at 100 ns. The blue trace represents the protein trajectory, while the red trace represents the ligand trajectory. (B) The RMSF plot of TNF. (C) The monitored interactions between proteins and ligands are categorized. (D) A schematic of detailed ligand atom interactions with the protein residues. (E) Timeline representation of protein-ligand interactions. The top panel illustrates the total number of specific contacts between the protein and the ligand over the simulation trajectory. The bottom panel indicates which residues interact with the ligand at each frame of the trajectory. Residues that form multiple specific contacts with the ligand are represented by a darker shade of orange, as per the scale provided to the right of the plot.
Effect of RA on HNEpCs proliferation
The HNEpCs were treated with varying concentrations of lyophilized powder of RA. The results (Fig. 6A) demonstrated that RA intervention at concentrations of 0.16 mg/mL, 0.3125 mg/mL, 0.625 mg/mL, 1.25 mg/mL, 2.5 mg/mL, 5 mg/mL and 10 mg/mL for a duration of 24 h significantly enhanced cell proliferation compared to the control group (0 mg/mL). However, the group treated with a concentration of 20 mg/ml exhibited inhibitory effects on cell growth without statistical significance. Additionally, after RA intervention period of 48 h, concentrations of 0.625 mg/mL, 1.25 mg/mL and 2.5 mg/mL showed significant promotion in cell proliferation, while concentrations of 0.16 mg/mL, 0.3125 mg/mL and 10 mg/mL had no effect on cell growth. The group treated with a concentration of 20 mg/ mL of RA displayed statistically significant inhibition of cells. The concentration of 5 mg/mL was ultimately chosen for cell intervention.
The effect of RA on protein levels of main targets of AR
The HNEpCs were exposed to control conditions, IL-13, and IL-13 plus RA for 24 h. The protein levels of STAT3, pSTAT3, NF-κB, and pNF-κB were quantified using Western blot (WB) analysis. Compared to the control group, the IL-13-induced inflammatory cell model group showed increased levels of pNF-κB, STAT3, and pSTAT3, while the levels of NF-κB protein remained unaffected. Additionally, treatment with RA inhibited the levels of pNF-κB, STAT3, and pSTAT3 proteins, as depicted in Fig. 6B.
The impact of RA on mRNA expression levels of main targets of AR
The HNEpCs were exposed to various treatments: control, IL-13 alone and IL-13 in combination with RA, for durations of 12 and 24 h. The mRNA expression levels of target genes were quantified using RT-qPCR. The PCR results (Fig. 7) revealed a significant increase in the mRNA expression levels of CCL2, IL-1β, IL-6, NF-κB, SP1 and HIF1A in the IL-13 group compared to those in the control group; However, IKBKB was found to be downregulated. Furthermore, as compared to the control group, there was no statistically significant difference in the expression of TNF-α and CHUK after 12 h of therapy or NFKBIA, CHUK, and CXCL8 after 24 h of treatment. In contrast to the IL-13 group, the expression levels of CCL2, IL-1β, IL-6, NF-κB, SP1, SMAD3 and HIF1A were significantly reduced in the IL-13 plus RA group; meanwhile, IKBKB was upregulated. Furthermore, no statistically significant difference was found in TNF-α expression after 12 h or NFKBIA expression after both 12 and 24 h between groups. Surprisingly, however, the gene expression level of CXCL8 increased in the RA group compared to the control and IL-13 groups.
Discussion
RA is frequently prescribed for the treatment of AR and has exhibited excellent efficacy in TCM. Studies have shown that RA reduces the number of eosinophils in the nasal mucosa and decreases the level of inflammatory cells in nasal lavage fluid while decreasing serum levels of IgE, IL-4, IL-5, and IL-13 and increasing IFN-γ and IL-10 levels20. Furthermore, RA reduced the secretion of inflammatory cytokines by increasing the ratio of CD4+ CD25+ Foxp3+ T cells and inhibiting the activation of nuclear factor-κB (NF-κB)20. However, the therapeutic targets and active ingredients of RA against AR have not been identified. This research utilized system pharmacology and bioinformatics analyses to screen targets, biological functions, and mechanisms. It has discovered potential key biotargets and molecular pathways of RA against AR, and then the above results were verified by in vitro experiments.
Firstly, we predicted the physicochemical properties and biological activities of the active ingredients of RA, including drug-likeness, absorption, distribution, metabolism, and excretion (ADME), which were assessed based on Lipinski’s Rule of Five, TPSA, and %ABS. These findings indicated that most key active phytochemicals fit the criteria of MilogP < 5, MW < 500 Da, HBA < 10, and HBD < 5 without violating the simplified Lipinski’s rule of five except for FA, 5’-hydroxyiso-muronulatol-2’, 5’-di-O-glucoside and isomucronulatol-7,2’-di-O-glucosiole. Furthermore, the main active components with TPSA < 140 Å2 and %ABS > 63% (aside from 9,10-dimethoxypterocarpan-3-O-β-D-glucoside and the two previously mentioned substances) fell within the optimal range for oral bioavailability. The analysis revealed that RA is theoretically free of drug-likeness and ADME restrictions. Additionally, molecules with bioactivity scores exceeding 0.00 are expected to show significant biological activity, whereas those with scores between − 5.0 and 0.00 show moderate activity, and those with scores below − 5.0 show inactivity21. Bioactivity analyses indicate that the pharmacological effects of RA involve multiple mechanisms, including ion channel modulators, enzyme inhibitors, protease inhibitors, GPCR ligands, nuclear receptor ligands, and kinase inhibitors. Thus, the pivotal active phytochemicals of RA are synergistic in treating disease.
Secondly, bioinformatics techniques and in vitro experiments were employed to ascertain potential targets. We identified 747 potential targets of RA against AR, and 15 targets were screened as main therapeutic targets in the PPI network. Molecular docking revealed that the active phytochemicals had a strong ability to bind to the primary targets, and molecular dynamics confirmed that the major complexes were stable. Combining the results of network pharmacology prediction and experimental validation, we discovered that RA could mitigate IL-13-induced inflammation by acting on TNF, NFKB1, IKBKB, CHUK, HIF1A, SP1, CCL2, IL-6, SMAD3 and IL-1β.
TNF stimulates proinflammatory and stress-response signals in several types of cell types22. Recent research has shown that susceptibility to AR is associated with tumor necrosis factor-α (TNF-α) gene polymorphisms23. Allergen-induced inflammation triggers a TNF-dependent innate memory, which could sustain and aggravate chronic type 2 airway inflammation24. Reports have also shown that TNF-α/NF-κB pathway inhibition might effectively reduce allergic airway inflammation25. NFKB1 is the transcription factor with the highest expression in macrophages, and it is also a critical cellular driver of inflammation and immunity26. Previous research has shown that M2 macrophages are associated with the severity of AR symptoms and enhance type 2 inflammation27. NFKB1 deletion intensifies canonical NF-κB signaling and boosts the content of macrophages28. IKBKB, also known as IKKβ/IKK2, involves in mediating the classical NF-κB pathway29. Deficits in IKBKB cause an impaired response to activation stimuli in a variety of immune cells, resulting in clinically relevant impairments in adaptive and innate immunity30. CHUK, a catalytic subunit of IκB kinase, has been reported to suppress the activity of the transcription factor NF-κB through phosphorylating the regulatory molecule TAX1BP131. The reduction in CHUK, which encodes IKKα, promotes inflammation related to NF-κB targets in pancreatic cells32. Therefore, CHUK plays a crucial role in the negative feedback of NF-kappa-B canonical signaling to restrict the activation of inflammatory genes. HIF1A plays a pivotal role as a transcription factor from a mechanistic standpoint. Pharmacologically or genetically, inhibiting HIF1A can mitigate macrophage inflammation induced by inf-exo-triggered hyperglycolysis. Additionally, administering an HIF1A inhibitor in vivo can alleviate experimental osteoarthritis33. SP1 is a transcription factor that plays a crucial role in regulating gene expression. It can activate or repress transcription in response to various stimuli. SP1 binds to specific DNA sequences known as GC-rich motifs and controls the expression of a broad array of genes involved in essential cellular processes such as cell growth, apoptosis, differentiation and immune responses. Knockdown of SP1 suppresses M1 polarization and related inflammatory responses in microglia by inhibiting the NF-κB signaling pathway. Moreover, silencing SP1 inhibited the activation of microglia and their mediated inflammatory responses34. The role of C-C chemokine ligand 2 (CCL2) and its corresponding receptor CCR2 in various inflammatory conditions has been extensively studied. Evidence suggests that the CCL2/CCR2 pathway extends to immune cell recruitment, inflammation and even metabolic syndrome35. Previous study has indicated that circ-Phkb enhances LPS-induced alveolar macrophage apoptosis and inflammation through the TLR4/MyD88/NF-κB/CCL2 axis36. IL-6 belongs to the pro-inflammatory cytokine family, stimulating the expression of diverse proteins implicated in acute inflammation and pivotal in human cell proliferation and differentiation37. Previous research has shown that within the IL-6 amplifier, stimulation of the IL-6-STAT3 and NF-κB pathways leads to an excessive production of IL-6, chemokines and growth factors, fostering the development of chronic inflammation that precedes the onset of inflammatory diseases38. SMAD3, a pivotal signaling transduction protein, is inhibited by miR-146a-5p. ESP-B4 facilitates the secretion of miR-146a-5p, modulates the Th1/Th2 balance of helper T cells, and alleviates symptoms of AR through Smad3/GATA-3 interaction39. IL-1β is a potent pro-inflammatory cytokine. Fibroblast-macrophage cell circuits were essential in the progression of airway inflammation and remodeling via IL-1β paracrine signaling40. Research has showed that excessive release of bioactive IL-1β may contribute to severe persistent AR inflammation, suggesting IL-1β as a biomarker for allergic diseases such as AR, atopy and asthma41.
Finally, we preliminarily validated the potential key mechanisms of RA in the treatment of AR. The enrichment analysis revealed that the crucial targets of RA are predominantly involved in inflammatory and immune responses, responses to lipopolysaccharides, cytokine production, and epithelial cell proliferation. They mainly exert their effects through the NF-κB signaling pathway, the TNF signaling pathway, the IL-17 signaling pathway, the Toll-like receptor signaling pathway, and the NOD-like receptor signaling pathway. Further in vitro experiments verified the inhibitory effect of RA on the expression levels of pNF-κB, STAT3, and pSTAT3 in the inflammatory HNEpCs model. This suggests that one of the possible mechanisms by which RA counteracts AR is through the regulation of the NF-κB/STAT3 signaling pathway.
The NF-κB signaling pathway is considered an archetypal proinflammatory signaling pathway, mainly due to the role of NF-κB in the expression of proinflammatory genes such as cytokines, chemokines and adhesion molecules42. Suppressing NF-κB signaling pathway may assist in controlling allergic responses in AR43. Moreover, RA and its main ingredients, formononetin, have been demonstrated in experiments to inhibit activation of the NF-κB signaling pathway and production of the inflammatory factors TNF-α and IL-1β44. RA exerts its anti-allergic effect through multiple pathways, including the NF-κB signaling pathway, while also being able to comprehensively modulate these aforementioned therapeutic targets to counteract the occurrence of AR.
The strength of our study is integrating of network pharmacological analyses from multiple databases to identify comprehensive potential therapeutic targets and mechanisms of RA against AR. The validation of the potential targets through molecular docking, dynamics simulations and in vitro experiments presents a more comprehensive screening and validation approach. To our knowledge, this research is the first time to provide a comprehensive insight into the potential therapeutic targets of RA against AR, which offering important strategies for its clinical application.
Nonetheless, there are limitations to the study. First, the validation of identified targets was solely based on computer simulations and in vitro assays, without in vivo confirmation. Secondly, our findings did not establish an optimal therapeutic dosage of RA for AR in clinical settings. Consequently, in vivo experimental and pharmacological studies are necessary in depth to better guide clinical practice. We will also add refinements to provide more favorable evidence in subsequent experiments.
Conclusion
Through network pharmacology analysis and verification by cell experiments, we have discovered and validated the potential key targets and mechanisms of RA against AR. The study showed that RA treatment may reverse the abnormal target expression, which is related to inflammation and immune regulation. The research also suggests that the key phytochemicals of RA have superior drug likeness and bioactivity, and the mechanism of RA against AR is likely the result of direct or indirect synergistic effects via multiple targets and pathways.
Data availability
The original data presented in the study are included in the article, and further inquiries can be directed to the corresponding authors.
References
Bousquet, J. et al. Allergic rhinitis. Nat. Rev. Dis. Primers. 6, 95 (2020).
Licari, A. et al. Epidemiology of allergic rhinitis in children: a systematic review and Meta-analysis. J. Allergy Clin. Immunol. Pract. 11, 2547–2556 (2023).
Vandenplas, O. et al. Impact of Rhinitis on Work Productivity: a systematic review. J. Allergy Clin. Immunol. Pract. 6, 1274–1286e9 (2018).
Cheng, L. et al. Chinese Society of Allergy Guidelines for Diagnosis and treatment of allergic Rhinitis. Allergy Asthma Immunol. Res. 10, 300–353 (2018).
Bousquet, J. et al. The allergic Rhinitis and its impact on Asthma (ARIA) Approach of Value-added Medicines: As-Needed treatment in allergic Rhinitis. J. Allergy Clin. Immunol. Pract. 10, 2878–2888 (2022).
Chen, Z. et al. Astragali Radix (Huangqi): a promising edible immunomodulatory herbal medicine. J. Ethnopharmacol. 258, 112895 (2020).
P, G. et al. Anti-aging function and molecular mechanism of Radix Astragali and Radix Astragali preparata via network pharmacology and PI3K/Akt signaling pathway. Phytomedicine: Int. J. Phytotherapy Phytopharmacology 84, (2021).
Zhang, C. et al. Database mining and animal experiment-based validation of the efficacy and mechanism of Radix Astragali (Huangqi) and Rhizoma Atractylodis Macrocephalae (Baizhu) as core drugs of traditional Chinese medicine in cancer-related fatigue. J. Ethnopharmacol. 285, 114892 (2022).
Guo, Z. et al. A systematic review of Phytochemistry, Pharmacology and Pharmacokinetics on Astragali Radix: implications for Astragali Radix as a Personalized Medicine. Int. J. Mol. Sci. 20, 1463 (2019).
Huang, J., Chen, X. & Xie, A. Formononetin ameliorates IL–13–induced inflammation and mucus formation in human nasal epithelial cells by activating the SIRT1/Nrf2 signaling pathway. Mol. Med. Rep. 24, 832 (2021).
Oh, H. A., Han, N. R., Kim, M. J., Kim, H. M. & Jeong, H. J. Evaluation of the effect of kaempferol in a murine allergic rhinitis model. Eur. J. Pharmacol. 718, 48–56 (2013).
Lipinski, C. A., Lombardo, F., Dominy, B. W. & Feeney, P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv Rev. 46, 3–26 (2001).
Bader, G. D. & Hogue, C. W. V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 4, 2 (2003).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa, M., Furumichi, M., Sato, Y. & Kawashima, M. Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592 (2023).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Gu, Z., Hübschmann, D. & simplifyEnrichment A Bioconductor Package for Clustering and Visualizing Functional Enrichment results. Genomics Proteom. Bioinf. 21, 190–202 (2023).
Rashid, M. Design, synthesis and ADMET prediction of bis-benzimidazole as anticancer agent. Bioorg. Chem. 96, 103576 (2020).
Gaillard, T. Evaluation of AutoDock and AutoDock Vina on the CASF-2013 Benchmark. J. Chem. Inf. Model. 58, 1697–1706 (2018).
Bing, Z. et al. Effect of Astragalus Membranaceus in Ovalbumin-Induced allergic Rhinitis Mouse Model. Am. J. Rhinol Allergy. 33, 420–432 (2019).
Hassan, M., Ashraf, Z., Abbas, Q., Raza, H. & Seo, S. Y. Exploration of Novel Human tyrosinase inhibitors by Molecular modeling, Docking and Simulation studies. Interdiscip Sci. 10, 68–80 (2018).
Aggarwal, B. B. Signalling pathways of the TNF superfamily: a double-edged sword. Nat. Rev. Immunol. 3, 745–756 (2003).
Cui, Q., Li, J. & Wang, J. The Assessment of TNF-α Gene Polymorphism Association with the risk of allergic Rhinitis in the Chinese Han Population. Int. J. Gen. Med. 14, 5183–5192 (2021).
Lechner, A. et al. Macrophages acquire a TNF-dependent inflammatory memory in allergic asthma. J. Allergy Clin. Immunol. 149, 2078–2090 (2022).
Xue, K. et al. Panax notoginseng saponin R1 modulates TNF-α/NF-κB signaling and attenuates allergic airway inflammation in asthma. Int. Immunopharmacol. 88, 106860 (2020).
Somma, D., Kok, F. O., Kerrigan, D., Wells, C. A. & Carmody, R. J. Defining the role of Nuclear factor (NF)-κB p105 subunit in human macrophage by Transcriptomic Analysis of NFKB1 knockout THP1 cells. Front. Immunol. 12, 669906 (2021).
Lou, H. et al. M2 macrophages correlated with symptom severity and promote type 2 inflammation in allergic rhinitis. Allergy 74, 2255–2257 (2019).
Best, K. T., Lee, F. K., Knapp, E., Awad, H. A. & Loiselle, A. E. Deletion of NFKB1 enhances canonical NF-κB signaling and increases macrophage and myofibroblast content during tendon healing. Sci. Rep. 9, 10926 (2019).
Gong, Y. et al. IKBKB rs2272736 is Associated with gastric Cancer survival. Pharmgenomics Pers. Med. 13, 345–352 (2020).
Pannicke, U. et al. Deficiency of innate and acquired immunity caused by an IKBKB mutation. N Engl. J. Med. 369, 2504–2514 (2013).
Deng, L. et al. Increase in IkappaB kinase alpha expression suppresses the tumor progression and improves the prognosis for nasopharyngeal carcinoma. Mol. Carcinog. 54, 156–165 (2015).
Li, X. & Hu, Y. Attribution of NF-κB activity to CHUK/IKKα-Involved carcinogenesis. Cancers (Basel). 13, 1411 (2021).
Liu, B. et al. Inflammatory fibroblast-like Synoviocyte-Derived exosomes aggravate osteoarthritis via enhancing macrophage glycolysis. Adv. Sci. (Weinh). e2307338 https://doi.org/10.1002/advs.202307338 (2024).
Guo, H. et al. Sp1 regulates the M1 polarization of Microglia through the HuR/NF-κB Axis after spinal cord Injury. Neuroscience 544, 50–63 (2024).
Wu, Y. & Ma, Y. CCL2-CCR2 signaling axis in obesity and metabolic diseases. J. Cell. Physiol. https://doi.org/10.1002/jcp.31192 (2024).
Wei, X. et al. Circ-phkb promotes cell apoptosis and inflammation in LPS-induced alveolar macrophages via the TLR4/MyD88/NF-kB/CCL2 axis. Respir Res. 25, 62 (2024).
Uciechowski, P. & Dempke, W. C. M. Interleukin-6: a masterplayer in the Cytokine Network. Oncology 98, 131–137 (2020).
Naito, S. et al. DDX6 is involved in the pathogenesis of inflammatory diseases via NF-κB activation. Biochem. Biophys. Res. Commun. 703, 149666 (2024).
Li, H., Wang, Y. & Han, X. ESP-B4 promotes nasal epithelial cell-derived extracellular vesicles containing miR-146a-5p to modulate Smad3/GATA-3 thus relieving allergic rhinitis: ESP-B4/miR-146a-5p in AR. Phytomedicine 108, 154516 (2023).
Yu, X. et al. Single-cell analysis reveals alterations in cellular composition and cell-cell communication associated with airway inflammation and remodeling in asthma. Respir Res. 25, 76 (2024).
Han, M. W., Kim, S. H., Oh, I., Kim, Y. H. & Lee, J. Serum IL-1β can be a biomarker in children with severe persistent allergic rhinitis. Allergy Asthma Clin. Immunol. 15, 58 (2019).
Sun, S. C. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 17, 545–558 (2017).
Wang, J. et al. MiR-146a mimic attenuates murine allergic rhinitis by downregulating TLR4/TRAF6/NF-κB pathway. Immunotherapy 11, 1095–1105 (2019).
K, X. et al. Formononetin protects LPS-Induced Mastitis through suppressing inflammation and enhancing blood-milk Barrier Integrity via AhR-Induced src inactivation. Front. Immunol. 13, (2022).
Acknowledgements
We need to send our appreciation to Yuanming Yang for his guidance in network pharmacology and molecular docking.
Funding
This research was supported by the National Natural Science Foundation of China (No. 82305328, 82274591, 82205204), the Guangdong Basic and Applied Basic Research Foundation from the Department of Science and Technology of Guangdong Province (No. 2021A1515220007), the Traditional Chinese Medicine Bureau of Guangdong Province (Guangdong Chinese Medicine Office Letter No. 52 [2022]), and the Science and Technology Program of Guangzhou from the Guangzhou Municipal Science and Technology Bureau (No. 202201020473).
Author information
Authors and Affiliations
Contributions
Y. H. prepared and wrote the original manuscript. Y.H and X.T. performed the network pharmacology and experiments. Y.H.,X.T. , and J. Z. were responsible for data curation and reviewed the manuscript. Y. L., Q.L., and W. C. were responsible for the conceptualization, funding acquisition, and supervision. N.X, S. Z., K. L., S. L. and R. C. analyzed the data. All authors approved the manuscript for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.

Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hua, Y., Tan, X., Zhang, J. et al. Deciphering the pharmacological mechanism of Radix astragali for allergic rhinitis through network pharmacology and experimental validation. Sci Rep 14, 29873 (2024). https://doi.org/10.1038/s41598-024-80101-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-80101-1
Keywords
This article is cited by
-
Exploration of the antitumor effect and mechanism of gambogic acid on osteosarcoma through network pharmacology and experimental pharmacology study
Scientific Reports (2025)
-
Mechanistic insights into Sanbi Decoction for osteoarthritis treatment based on network pharmacology and experimental validation
Scientific Reports (2025)