Abstract
Understanding the genetic basis of drought tolerance in safflower (Carthamus tinctorius L.) is essential for developing resilient varieties. In this study, we performed a genome-wide association study (GWAS) using DArTseq markers to identify marker-trait associations (MTAs) linked to drought tolerance across 90 globally diverse safflower genotypes. These genotypes were evaluated under both rainfed (drought) and irrigated conditions over three consecutive years (2016–2018). Significant variation in seed yield and morphological traits was observed between genotypes and across environments, with notably reduced performance under rainfed conditions. A total of 66 MTAs were identified for key agronomic traits, including seed yield, 1000-seed weight, number of heads per plant, and days to maturity, with 45 MTAs showing significant associations with the stress-tolerance index (STI). Twelve MTAs were consistently detected across both stress and non-stress conditions, suggesting stable loci for breeding applications. BLAST searches indicated that several identified markers corresponded to genes involved in abiotic stress response, including biotin carboxylase, serine/threonine-protein kinase, and zinc finger domain proteins. The clustering of genotypes based on drought-tolerance coefficient (DC) and STI values grouped genotypes into drought-susceptible and drought-tolerant categories, with no strong correlation to geographic origin. These findings provide valuable insights into the genetic architecture of drought tolerance in safflower and highlight candidate loci for marker-assisted selection, supporting the development of drought-resilient varieties.
Similar content being viewed by others
Introduction
Safflower (Carthamus tinctorius L.) is a significant oilseed crop valued for its edible oil, natural dye, and a high-quality use in poultry feed1,2. Iran is among the largest importers of oilseeds, cooking oil production and as components of animal feed. In recent years, increasing demand for oilseed crops has intensified interest in the cultivation and breeding of adaptable oilseed crops such as safflower3,4. Renowned for its resilience to drought and salinity, safflower is believed to have been domesticated over 4000 years ago in the Fertile Crescent (encompassing present-day Israel, Syria, Iran, Iraq, and Turkey) and the Indian subcontinent5,6,7. Water-deficit stress is one of the most significant abiotic stresses due to its unpredicted timing, severity, and duration. Many safflower-producing countries, including India, Pakistan, Turkey, and Iran, are located in semi-arid regions characterized by winter-dominant rainfall. Consequently, safflower plantations in these areas often suffer yield losses caused by terminal drought stress8,9. While safflower exhibits notable tolerance to water stress, its seed yield is negatively affected by terminal drought during the flowering and seed-filling stages, similar to other crops10,11.
Although safflower exhibits the ability to tolerate drought stress with only a slight reduction in seed yield, it is considered a climate-resilient crop. However, research efforts and genetic insights into safflower’s drought tolerance mechanisms remain limited compared to other oilseed crops, likely due to its restricted cultivation area status as a minor crop worldwide12,13. Improving seed yield under stress conditions is more challenging than in favorable environments, as seed yield is a complex trait governed by multiple genes and significantly influenced by environmental factors14. Therefore, understanding the genetic regulation of seed yield, oil content, and morphological traits in safflower is crucial. This knowledge can enable breeders to design effective strategies for developing high-yield, high-oil content safflower genotypes that are drought-tolerant in arid regions11.
Drought stress tolerance in plants is a highly complex trait regulated by numerous genes, making it challenging to directly select drought-tolerant genotypes based on a single trait. The drought-tolerance coefficient (DC) and stress-tolerance index (STI) quantify trait loss under drought stress compared to non-stress conditions. These indices are widely used across various crop species as reliable measures for the indirect selection of drought-tolerant genotypes15,16.
An essential breeding strategy for developing improved genotypes involves identifying genes or quantitative trait loci (QTL) associated with seed yield and key agronomic traits through multi-environmental phenotypic and high-throughput molecular analyses15. Various molecular markers, such as SSR, AFLP, RAPD, and SNP have been employed to assess genome diversity in safflower17,18. However, many of these studies face limitations, including low genome coverage, low polymorphism, high costs and time-consuming processes. Advances in next-generation sequencing (NGS) techniques now enable researchers to generate thousands of loci with extensive genomes coverage using a DNA chip. These advancements have significantly reduced cost and improved efficiency for whole-genome sequencing (WGS) techniques across crop species, including safflower18,19,20.
In this context, to reduce the cost and time required for generating markers with wide genome coverage, a microarray-based technology called Diversity Array Technology (DArT) developed a genotyping-by-sequencing (GBS) platform known as DArTseq for various plant species21,22,23 , including safflower7,24. An additional advantage of DArTseq technology is its superior genome coverage compared to other GBS techniques, due to its higher sequencing depths and strict marker filtering criteria25.
While characterizing genes or QTLs associated with desirable traits in plants using bi-parental mapping populations, several limitations arise. This include low recombination resulting from the limited alleles contributed by each parent and small mapping populations, both of which lead to low mapping resolution26. Compared to traditional bi-parental QTL mapping, genome-wide association studies (GWAS) offer higher mapping resolution by analyzing a larger number of loci in a diverse set of individuals from the germplasm. This approach generates broader genetic variation and allows the simultaneous evaluation of many alleles across the germplasm, making it faster and more cost-effective, especially for complex quantitative traits27,28. To our knowledge, only a few genome-wide association studies (GWAS) have been conducted on safflower for genetic dissection of agronomic traits. These studies have often been limited by a small number of markers or have focused exclusively on traits evaluated under rainfed conditions24,29.
Building on this background, this study evaluated a panel of geographically diverse safflower genotypes in field conditions under two water regimes (irrigated and rainfed) over three years (2016–2018). Additionally, a genome-wide association mapping analysis was performed using high-density chromosome-level assembled DArTseq markers to identify QTLs associated with seed yield, seed oil content, and agronomic traits across both environments. Notably, this research represents the first report of chromosome-level positioning of DArTseq markers based on the safflower reference genome.
Methods
Plant materials and field assessment
A panel of 90 safflower genotypes, preserved at Iranian Seed and Plant Improvement Institute (SPII), Iran was used in this study. These genotypes was selected based on a preliminary molecular phylogenetic analysis and the agronomic performance of 135 genotypes4. The panel represents germplasm from 12 countries, including regions of origin and domestication center of safflower6,7. Detailed information on genotype code, name and origin is provided in Supplementary Table S1. The association panel was cultivated under both non-stressed and water deficient (rainfed) environments at the Dryland Agricultural Research Institute (DARI) of Sararood, Kermanshah, Iran (47° 20ʹ N latitude, 34° 20ʹ E longitude, and 1351.6 m altitude) across three consecutive seasons (2016, 2017 and 2018). Kermanshah features a hot Mediterranean/dry-summer subtropical climate (Köppen-Geiger classification: Csa), characterized by mild temperatures with moderate seasonal variation. Summers are typically dry and hot due to subtropical high-pressure systems, while winters are marked by moderate temperatures and variable, rainy conditions influenced by the polar front.
Detailed information on soil physicochemical properties, as well as monthly precipitation and temperature data (2016–2018), is provided in Supplementary Table S2. The field experiment followed a 10 × 9 lattice design with two replications per irrigation regime (rainfed and irrigated) each year. Seeds were planted in three rows of 3 m per plot, with 50 cm spacing between rows and 20 cm spacing between plants within rows. Sowing occurred on March 1st, and harvesting was completed by September 10th for all experiments and environments. Water-deficit stress plots were maintained under rainfed conditions (no irrigation), while non-stress plots received three irrigation at 10-day intervals from the flowering to seed-filling stages. Favorable rainfall and temperature during March and April supported germination and plant establishment in the field. However, very low rainfall and high temperature during May to September (flowering to maturity stages) created a "drought stress environment" for the rainfed treatments in this experiment.
Morphological data collection and statistical analysis
Phenotypic traits for each genotype were assessed based on ten plants from the middle row in each replication across both environments. At maturity, data were recorded for seed yield (SY), 1000-seed weight (TSW), number of heads per plant (NHP), plant height (PH), head diameter (HD), and number of seeds per head (NSH). Days to flowering (DTF) were recorded when 50% of plants in each plot had flowered, while day to maturity (DTM) were recorded when 50% of the plants reached maturity. Seed oil content (OIL) was measured using 10 g of seeds from each genotype in both environments using the Soxhlet method and calibration of NIRS equations for safflower oil content12,30. Relative-water content (RWC) for each genotype in both environments was calculated using the following equation31:
Here, LFW represents the leaf fresh weight, LDW is the leaf dry weight after drying at 72 °C for 5 days, and LTW is the turgid weight of leaves after soaking in water for 6 h.
The three-year data sets in both environments were initially analyzed separately for each trait using a linear mixed model (LMM) that accounted for spatial effects (row and column effects). Analysis of variance (ANOVA) was performed to evaluate the effects of genotype, year, irrigation regimes and their interactions, using SPSS software (IBM Inc., Armonk, NY, United States). The resulting best linear unbiased estimator (BLUE) values for each genotype were then used to fit a multiple-environment (years) LMM, treating years as random effects32. The BLUE values for each quantitative trait were subsequently used for statistic descriptive parameters and Pearson’s correlation coefficient analysis between measured traits, utilizing the “cor” function in the R platform. Additionally, the “FactoMineR” package was employed to conduct principal component analysis (PCA) on the correlation matrix to examine the distribution of quantitative traits.
To assess the adaptation of safflower genotypes to drought stress, the BLUE values for each trait under drought stress (DS) and non-stress (NS) conditions were used to calculate the drought-tolerance coefficient (DC) using the following equation15:
The drought-tolerance stress index (STI) was then calculated using the following formula33:
where, YDS is the performance of a genotype in the DS environment, YNS is the performance of the same genotype in the NS environment and \({{\bar{Y}}}_{NS}\) is the mean YNS of all genotypes. The DC and STI values summarized in supplementary Table S3. Based on measured morphological traits, as well as STI and DC-values, safflower genotypes were grouped by calculating Euclidean distance and applying the weighted pair group method with arithmetic mean (WPGMA) to evaluate their drought tolerance level.
DArTseq markers, filtering and genome assembly
Ten seeds from each safflower genotype were sown in pots, and after 20 days, fresh pooled leaves from each pot were used for DNA extraction using the CTAB protocol34, with minor modification35. The quality and quantity of extracted DNA were assessed via 0.8% agarose gel electrophoresis, and the final concentration was adjusted to 100 ng/µl. DNA samples from 90 safflower genotypes were sent to Diversity Arrays Technology Pty Ltd, Canberra, Australia (https://www.diversityarrays.com/), for genotyping by sequencing (GBS) analysis using high density SilicoDArT and SNP markers, as previously described for safflower7 and other crop species36,37,38. Quality parameters, including call rate, polymorphic information content (PIC), and marker reproducibility, were computed using DArTsoft v.7.4.7 (provided by DArT Pty Ltd). Markers were filtered to retain those with a minor allele frequency (MAF) of at least 5% and a maximum of 10% missing data24,39. This study reports the DArTseq markers for these safflower genotypes for the first time. Since their chromosomal locations were previously unknown, the physical position of the filtered markers were determined by mapping their flanking sequence to the draft safflower genome assembly (http://safflower.scuec.edu.cn), which includes 12 pseudochromosomes and was recently reported20.
Genetic diversity, population structure and linkage disequilibrium (LD)
To evaluate the phylogenetic diversity among the safflower genotypes, an identity-by-state (IBS) distant matrix was calculated in TASSEL v.5.2.3740. Cluster analysis based on this genetic distance matrix was performed using the unweighted neighbor joining (UNJ) algorithm, while principal coordinate analysis (PCoA) was conducted in DARWIN ver. 5.0 software41. Population structure analysis utilized a Bayesian clustering approach implemented in Structure 2.142, with five independent runs. Each run consisted of 50,000 burn-in iterations followed by 50,000 Markov Chain Monte Carlo (MCMC) iterations for each KKK value (number of populations), ranging from 2 to 10. The optimal number of populations was determined using Structure Harvester (web version 0.6.94)43. Analysis of molecular variance (AMOVA) was conducted to assess genetic variation within and between the estimated subpopulations (K) using GeneAlex Excel add-in software v.6.544.
Linkage disequilibrium analysis was performed using the GAPIT package within the R software environment. Squared allele frequency correlations (r2) with p-values < 0.001 were used to identify significant LD between locus pair. The LD pattern was visualized with a heatmap generated using the LDHeatmap package in R45. Additionally, a DArTseq marker density plot was created using the R package "CMplot".
Genome-wide association study (GWAS)
Genome-wide association studies (GWAS) were conducted using the Genomic Association and Prediction Integrated Tool (GAPIT) in R46. Marker-trait association (MTA) analysis was performed using two single-locus methods, the general linear (GLM) and mixed linear (MLM) models, and three multi-locus GWAS models: FarcmCPU47, BLINK48 and MLMM49. Manhattan and QQ plots were generated using R packages, qqman version 0.1.431. The BLUE values for each trait across both environments, along with the stress-tolerance index (STI) for each trait, were used as phenotypic data for GWAS. Population structure and the kinship similarity matrix were incorporated into the analysis based on the requirements of each model. Significant MTAs were identified for markers that exceeded the false discovery rate threshold (P = 0.05), with a significance level of P ≤ 0.0001 (log10 P ≥ 4).The flanking sequences of significant MTAs (within 10 K upstream and downstream windows) were extracted from the reference genome of Carthamus tinctorius20. These sequences were analyzed using BLAST against the NCBI database to annotate potential candidate genes associated with traits of interest.
Results
Phenotypic variation of safflower genotypes
In this study, diverse safflower genotypes were evaluated for seed yield, oil content, and morphological traits across multi-year field trials (2016–2018) conducted under drought-stressed and non-stressed conditions. These trials aimed to investigate genotype responses to water-limited and irrigated environments, providing critical insights for breeding drought-tolerant safflower varieties. Analysis of variance (ANOVA) showed a significant influence of the growing year on all traits (Table 1), underscoring the impact of environmental factors such as rainfall patterns, temperature fluctuations, and soil moisture levels specific to each growing season. Furthermore, highly significant (p < 0.01) variation in seed yield and all other traits was observed among genotypes under both drought-stressed and non-stressed conditions, indicating that genetic differences play a key role in determining trait expression across water regimes.
The significant three-way interaction (p < 0.01) among genotype, water regime, and year highlights the complexity of genotype-by-environment (G × E) interactions, emphasizing how genotype performance is influenced only by water availability but also by specific environmental conditions across year. Descriptive statistics based on the best linear unbiased estimator (BLUE) values for seed yield, oil content, and other yield-related traits are presented in Table 2. Under irrigated conditions, seed yield ranged from 590.84 to 2043.18 kg/ha, with a mean of 1115.02 kg/ha, and exhibited relatively low heritability (0.14). In the rainfed (drought-stressed) environment, seed yield showed a significant reduction, ranging from 356.02 to 1165.33 kg/ha, with an average of 673.26 kg/ha and a slightly lower heritability (0.12). This reduction in seed yield under drought stress underscores the sensitivity of safflower genotypes to water limitation, aligning with findings from other drought-stressed crops where yield losses are attributed to insufficient water during critical growth stages. Additionally, significant differences among genotypes were observed for yield-related traits in both environments, with consistently lower values recorded under rainfed conditions.
Interestingly, seed oil exhibited an opposite trend compared to seed yield, showing higher mean value under rainfed conditions. Oil content ranged from 19.11 to 39.95% with an average of 29.49% under drought-stressed conditions, compared to a range of 21.61 to 36.69% and an average of 27.60% under irrigated conditions (Table 2). This increase in oil content under water stress suggests that certain genotypes may enhance oil accumulation as a stress response, a trait of potential value for breeding drought-resilient safflower varieties. The coefficient of variation (CV) and heritability estimates were generally higher for seed yield and related traits under irrigated conditions, indicating more stable trait expression in favorable environments. This stability, along with reduced environmental variation under irrigation, may enable more predictable selection outcomes for yield-related traits in well-watered conditions. Conversely, the increased variability under drought stress highlights the need to incorporate stress-specific traits or indices, such as the stress-tolerance index (STI), to effectively identify genotypes that combine high yield potential with resilience to water limitation. Overall, measured traits exhibited higher CVs in the irrigated environments compared to the rainfed environments, while heritability estimates were also greater under irrigation (Table 2). Drought-tolerance coefficient (DC) values for various traits revealed distinct distributions among genotypes (Supplementary Table S3). The mean DC value for measured traits in tested safflower genotypes are presented in Table 2. Traits such as number of heads per plant (NHP), relative water content (RWC), head diameter (HD), number of seeds per head (NSH), and 1000-seed weight (TSW), which had DC values below 0.50, were particularly sensitive to water stress.
Under irrigated conditions, seed yield (YP) showed positive correlation with the number of heads per plant (NHP), number of seeds per head (NSH), 1000-seed weight (TSW), and oil content (OIL). Additionally, a significant positive correlation was observed between OIL and both head diameter (HD) and NSH. While TSW was positively correlated with NHP, it exhibited a negative correlation with NSH (Fig. 1). In the supplementary-irrigation environment, seed yield (Ys) was positively correlated with TSW and NSH, whereas OIL displayed a significant positive correlation with HD and a significant negative correlation with plant height (PH) (Fig. 1). Principal component biplot analysis further illustrated the relationships among seed yield, oil content, and agro-morphological traits (Fig. 2). Under supplementary irrigation, the first principal component (PC1) accounted for 36.7% of the total variation, with NSH, PH, and HD as key contributors, while the second component (PC2) explained 21.2%, primarily influenced by seed yield (Yp), NHP, and TSW (Fig. 2). Similarly, in the rainfed environment, principal component analysis (PCA) revealed that PC1 and PC2 explained 29.4% and 16.2% of the variation, respectively. Key contributors to PC1 included seed yield (Ys), NHP, HD, days to maturity (DTM), and NSH (Fig. 2).
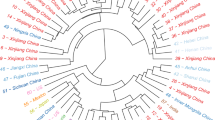
Cluster analysis based on the BLUEs values of measured traits under both irrigated and rainfed conditions grouped the 90 safflower genotypes into four and three clusters, respectively (Fig. 3). Under rainfed conditions, the genotypes were grouped into three clusters (Fig. 3a). Clusters I and II comprised 32 and 21 genotypes, respectively, which exhibited relatively higher seed yields compared to the 37 genotypes grouped into cluster III (Table 3). Under an irrigated conditions, the genotypes were divided into four distinct clusters (Fig. 3b). Cluster I and III contained 33 and 30 genotypes, respectively, which were characterized by low numbers of seeds per capitula and low 1000-seed weight, resulting in lower seed yields. However, these genotypes demonstrated higher seed oil content. In contrast, clusters II and IV comprising 24 and 3 genotypes, respectively, displayed high seed yield and related traits but relatively lower oil content (Table 3). Notably, the three genotypes in cluster IV were identified as superior genotypes for seed yield under irrigated conditions. Across both environments, there was no correlation between genotype clustering based on measured traits and their geographical origins.
To further classify the genotypes into drought-susceptible and drought-tolerant categories, cluster analysis was conducted using drought-tolerance coefficient (DC) and stress-tolerance index (STI) values. This analysis grouped the 90 safflower genotypes into four distinct clusters, each representing different levels of drought tolerance (Fig. 4).
Group I primarily consists of drought-susceptible genotypes, characterized by low drought-tolerance coefficient (DC) and stress-tolerance index (STI) values. These genotypes experience significant yield reductions under drought stress, indicating high sensitivity to water deficits. Group II includes genotypes with moderate drought tolerance. While these genotypes perform reasonably well under optimal conditions, their yield declines under drought stress, classifying them as susceptible but less severely impacted than those in Group I.
Group III represents moderately drought-tolerant genotypes, which maintain relatively stable performance across both drought-stressed and irrigated conditions. These genotypes have DC values near the threshold for tolerance, reflecting their intermediate resilience. Group IV comprises highly drought tolerance genotypes, demonstrating minimal yield loss under drought conditions. These genotypes exhibit high DC and STI values, indicating strong adaptability to water-limited environments. In Group IV, key yield-related traits such as seed yield, 1000-seed weight, and number of seeds per head shows less variation between environments, highlighting the ability of these genotypes to maintain yield stability despite water stress. Conversely, drought-susceptible genotypes in Groups I and II exhibit substantial yield reductions and lower DC values. These genotypes also have lower STI values, underscoring their limited capacity to adapt to drought stress. Traits such as head diameter and relative water content (RWC) tend to decline more sharply in these groups, further reflecting their vulnerability to water stress.
The high STI and DC values observed in Group IV suggest that drought tolerance in these genotypes may be linked to specific physiological adaptations, such as the ability to maintain relative water content (RWC), a key trait associated with drought resilience. Notably, genotypes G33, G52, G51, G13, and G18 stand out for their superior STI and DC values, indicating exceptional drought tolerance. These genotypes may possess unique genetic traits, such as efficient water use or mechanisms to reduce transpiration under stress, making them valuable candidates for drought-resilient breeding programs.
The absence of strong correlation between geographic diversity and phenotypic clustering based on DC and STI suggests that drought tolerance traits are not strictly tied to geographic origin. This finding is advantageous for breeding programs, as it allows the exploration of genetic diversity across regions without constraints imposed by origin-based clustering.
DArTseq marker distribution
DArTseq genotyping technology was utilized to generate SNP and SilicoDArT markers for a panel of 90 safflower genotypes. This process produced a total of 19,639 markers, comprising 10,130 SilicoDArTs and 9,509 SNPs. After applying quality control filters (i.e., MAF > 0.1 and missing data per genotype < 10%) and aligning the markers on the 12 pseudochromosomes of the draft safflower genome20 (Carthamus tinctorius reference genome assembly GCA_018320725.1), a total of 7029 markers (2193 SNP and 4836 SilicoDArT) were retained for furthers analysis (Supplementary Table S4). The number of DArTseq per pseudochromosome ranged from 447 on Chr1 to 739 on Chr5, with an average of 585.7 markers per pseudochromosome (Table 4). The chromosome size covered by DArTseq markers varied from 66.76 Mbp on Chr1 to 106.47 Mbp on Chr3, corresponding to an average of 65.59 markers per Mbp across the entire genome (Table 4).
Population structure, genetic diversity and LD
Kinship coefficient values, calculated based on the average dissimilarity between genotypes, ranged from 0.18 to 0.63, with an average value of 0.48. A heatmap illustrating the relationship among safflower genotypes revealed three distinct groups (Fig. 5a). Additionally, the population structure, inferred using on Bayesian model, is visualized at K = 3 (Fig. 5b,c). The three subpopulations contained 21.2% (POP1), 23.4% (POP2) and 55.5% (POP3) of the safflower genotypes, with expected heterozygosity (genetic divergence) value of 0.05, 0.31 and 0.21 for POP1, POP2 and POP3, respectively. Cluster analysis using the Unweighted Neighbor-Joining (UNJ) algorithm also grouped the population into three subpopulations, consistent with the results of population structure analysis. Cluster I contained 21 genotypes from various countries, while cluster II and III included 29 and 40 genotypes, respectively (Fig. 5d). Overall, there was no correlation between genotype clustering and geographic origin. However, in clusters II and III, genotypes from Iran and Turkey tended to cluster together, while those from India were grouped closely. Analysis of molecular variance (AMOVA) revealed that the majority of genetic variation (85.5%) occurred among genotypes within populations, with a smaller proportion (14.5) attributed to variation among groups (Table 5).
Linkage disequilibrium (LD) for DArTseq markers with a minor allele frequency (MAF) > 10% was measured using pairwise marker r2 across the genome. LD was observed to be extensive, encompassing 350,176 marker pairs, of which 36,752 (10%) intra-chromosomal pairs exhibited significant level (P < 0.001). The mean r2-value and critical r2 were 0.11 and 0.14, respectively (Supplementary Figure S1). LD decay was observed over a distance of 1.86 MB, indicating that SNPs within this range are likely to behave as a single inheritance block.
Marker–trait associations (MTAs)
A multi-model GWAS analysis, incorporating both single-locus (GLM and MLM) and multi-locus (MLMM, FarmCPU and BLINK) identified significant marker-trait associations (MTAs) for morphological traits and the stress tolerance index (STI) of each trait. Among these models, FarmCPU demonstrated high computation efficiency by effectively controlling false positive and mitigating their effect in GWAS. Consequently, GWAS results using the FarmCPU model, along with the BLUE values for each trait, were used to identify significant MTAs. The significant MTAs for morphological traits and STI values are summarized in Tables 6 and 7, respectively. All genomic positions for markers and genes referenced in this manuscript are based on the Carthamus tinctorius reference genome assembly GCA_018320725.1, previously reported by Wu et al.23.
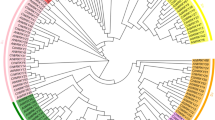
Under supplemented irrigation conditions, 31 significant MTAs (P ≤ 0.0001) were detected across all chromosomes except for chromosome 3 (Table 6). These significant MTAs are illustrated using Manhattan and QQ plots (Fig. 6; Supplementary Figure S2). These significant MTAs are illustrated using Manhattan plots and QQ plots (Fig. 6; Supplementary Figure S2). Chromosome 4 contained the highest number of significant MTAs (7), followed by chromosomes 1 and 10 (4 each), chromosomes 5, 6, and 8 (3 each), chromosomes 2 and 9 (2 each), and chromosomes 7, 11, and 12, each with one significant MTA. The highest number of significant MTAs were identified for plant height (PH), totaling six MTAs distributed across four chromosomes (4, 5, 6, and 10). Additionally, four MTAs (Cart-SNP212, Cart-SNP356, Cart-SNP3879, and Cart-SNP6353) were significantly associated with seed yield (SY) under irrigated conditions.
Circular Manhattan plot representing the markers-traits association for morphological traits in 90 safflower genotypes under supplementary irrigation environment. The trait indicators used for constructing the circular Manhattan plot from outside to inside are TSW, Yp, RWC, PH, OIL, NSH, NHP, HD, DTM and DTF, respectively.
The significant MTAs and their associated parameters for measured traits under rainfed conditions are summarized in Table 6. Under drought stress (rainfed condition), 35 significant MTAs (P ≤ 0.0001) were identified for 10 traits. The associations are illustrated using Manhattan and QQ plots (Fig. 7; Supplementary Figure S3). Significant MTAs were identified across various chromosomes, with the exception of chromosomes 7 and 12. The highest number of associations was observed on chromosomes 4, 6 and 10, each with six significant MTAs, followed by chromosome 5 with five MTAs (Table 6). Four MTAs (Cart-SNP212, Cart-SNP665, Cart-SNP1893 and Cart-SNP3025 ) showed significant association with seed yield (SY) under rainfed conditions. Only a limited number of MTAs were shared between irrigated and rainfed conditions, including DTM (Cart-SNP2139), NHP (Cart-SNP2113 and Cart-SNP3276), TSW (Cart-SNP4990) and SY (Cart-SNP212).
Circular Manhattan plot representing the markers-traits association for morphological traits in 90 safflower genotypes under rainfed environment. The trait indicators used for constructing the circular Manhattan plot from outside to inside are DTF, DTM, HD, NHP, NSH, OIL, PH, RWC, Ys and TSW, respectively.
Among the identified MTAs, one pleiotropic MTA (Cart-SNP2549) was detected under both conditions. This pleiotropic MTA, located on the chromosome 5, was associated with relative water content (RWC) and plant height (PH ) (Table 6).
A total of 45 MTAs were identified across the safflower genome for the analyzed STI values at a significance threshold of Log10p ≥ 4.0 (Table 7; Fig. 8). Significant MTAs were detected on all chromosomes. The highest number of MTAs was observed for STI-SY and STI-HD (6 each), followed by STI-TSW, STI-NHP and STI-DTM (5 each), STI-RWC, STI-PH and STI-DTF (4 each), STI-OIL and STI-NSH, with 3 significant MTA each (Table 7). Among these MTAs, 12 overlapped with MTAs identified for corresponding traits under drought-stress or non-stress conditions (Table 7), suggesting their stability. These stable markers were subsequently used for the identification of candidate genes. The locations of the significant MTAs were examined within 10 kb window on each side using the safflower reference genome (Carthamus tinctorius reference genome assembly GCA_018320725.1) to identify candidate genes associated with STI values. Gene ontology analysis for STI-DTF, STI-DTM, STI-HD, STI-NHP, STI-PH, STI-TSW and STI-SY traits identified 12 markers containing overlapping genes (Table 8). These MTAs were linked to several biological and molecular processes, including genes encoding biotin carboxylase, dehydrogenase gene, chlorophyll a-b binding protein, serine/threonine-protein kinase, zinc finger domain-containing protein, diacylglycerol acyl transferase gene and carboxylesterase.
Discussion
In Iran, Safflower is widely cultivated in dry and semi-dry regions, where it is typically sown in winter or early spring to maximize the use of available water for germination and vegetative growth. This study assessed drought tolerance in a diverse global collection of safflower genotypes under rainfed and supplementary irrigation conditions. Average monthly rainfall and temperature data recorded during the safflower growing seasons from 2016 to 2018 indicated wet condition in early spring, followed by a dry period from late spring to early summer. As a result, the experimental site experienced drought stress in May, coinciding with the flowering stage. The study revealed significant genotype-by-environment (G × E) interactions for all the traits analyzed. High G × E interactions for seed yield and morphological traits across different environments were consistent with findings from previous studies on safflower19,29.
Substantial differences were observed between the minimum and maximum values of seed yield and its component traits under rainfed and supplementary irrigation conditions, emphasizing the detrimental effects of water stress on all the measured traits. These findings align with previous studies on safflower subjected to water stress across various nvironments10,11,50,51. Understanding the heritability of traits and their underlying additive or non-additive gene actions offers valuable insights for successful trait transmission across generations. This knowledge is critical for designing effective breeding programs11,52.
The results of this study across both environments demonstrated high heritability for traits such as day to flowering (DTF), day to maturity (DTM) and thousand-seed weight (TSW) (Table 2), indicating strong genetic inheritance of these traits. Previous studies on safflower field evaluations in various environments have suggested that these highly heritable traits predominantly exhibit additive gene action. This additive effect likely plays a more significant role than non-additive effects in the genetic control of seed yield and its componentsincluding days to maturity, flowering time, and plant height under drought stress conditions11,53. consequently, selection strategies focused on these traits in safflower can be effective for developing high-yield, drought tolerance genotypes. Correlation coefficients between seed yield and its component traits were assessed in both environments. The results revealed a positive, though moderate, correlation (r2 = 0.37), between seed yield under stress and non-stress conditions. This finding suggests that selection solely on high yield performance under optimal conditions may inadvertently favor susceptible, low yield genotypes under drought stress. Previous studies in various crops have similarity shown that yield performance in optimal environments does not necessarily correlate with seed yield under drought stress conditions54,55.
Seed yield under non-stress conditions showed a positive correlation with number of heads per plant (NHP), number of seeds per head (NSH) and 1000-seed weight (TSW) (Fig. 1). Similarly, under water stress condition, seed yield was positively correlated with TSW and NSH, suggesting that selection safflower genotypes based on these traits could enhance yield under both water-stress and non-stress environments. Additionally, PCA-biplot analysis in both environments identified NHP, NSH, TSW and seed yield (Yp and Ys) as the most effective traits for assessing diversity and characterization safflower genotypes in this study. These finding are consistent with previous studies in safflower11,56.
Drought tolerance is a highly complex traits governed by multiple genes affecting seed yield and related characteristics. Consequently, direct yield and morphological traits alone may not suffice to identify drought tolerance genotypes. Drought tolerance indices, such as Stress Tolerance Index (STI )and Drought Coefficient (DC), are widely used in evaluation crops under abiotic stress conditions15,33. Successful breeding programs depend on selecting appropriate genotypes based on their trait performance and genetic potential. In this study, genotypes G33, G52, G51, G13 and G18, which exhibited high values for both STI and DC, were identified as superior for seed yield and positively associated traits such as TSW and NSH under both experimental conditions. These genotypes are considered stable and may possess unique genetic traits, such as efficient water use or mechanisms for reducing transpiration under stress, making them valuable candidates for breeding programs. The observed genotype-by-environment (G × E) interactions indicated that genotypes in Group IV (based on DC and STI values) likely harbor genetic mechanisms supporting enhanced phenotypic plasticity under varying environmental stresses. The classification of genotypes based on drought tolerance, as shown in Fig. 4, along with detailed trait data in Supplementary Table S3, provides a valuable resources for agronomists and breeders. Drought-tolerant genotypes demonstrating strong agronomic performance under both stress and non-stress conditions hold significant promise for sustainable production in water-scarce environments. This classification also offers insights into regional adaptability and helps inform resource allocation decisions for future breeding programs.
No significant correlation was observed between the clustering of genotypes based on drought stress values and the genomic relationships derived from DArTseq data. However, genotypes from specific clusters (e.g., those grouped by drought tolerance or susceptibility) may exhibit patterns of genetic similarity within the broader population structure or subgroups identified by the Bayesian clustering model (K = 3).
Genome-wide association studies (GWAS) are powerful and efficient tools for dissecting the genetic architecture of complex traits, such as seed yield and agronomic characteristics, under diverse environmental conditions in safflower19,29 and other crops. In this study, GWAS analysis were conducted to identify both environmentally stable and environmental-specific MTAs, providing valuable insights into the genetic basis of important traits and their genotype-by-environment (G × E) interactions29,57. We employed two single locus models (GLM and MLM) and three multi-locus models (MMLM, FarmCPU and BLINK), all of which are recognized for their reliability in GWAS analyses. Additionally, DArTseq markers, known for their high genome coverage and resolution, were used to analyze complex traits across various plant species effectively36,38.
The mean call rate (92%) and reproducibility (99%) of the DArTseq markers used in this study are consistent with previous reports, demonstrating the high quality and reliability of this marker set for association mapping. GWAS was conducted on 90 globally diverse safflower genotypes using 7029 DArTseq markers, which were mapped on 12 pseudochromosomes based on alignment with the safflower reference genome20.
At a significant level of p < 0.0001, a total of 31 and 35 MTAs were identified as associated with traits under irrigated and rainfed conditions, respectively (Table 6). In line with many studies on various plants species, the present study found that drought-stressed environments often yield more MTAs than irrigated environments, likely due to the stronger influence of environmental factors on genotype expression. For all traits, the majority of MTAs were specific to a single environment, suggesting that these site-specific MTAs are linked to high G × E interactions. This information can help breeders understand key traits interactions and guide targeted breeding efforts in specific environments29. These finding are consistent with previous reports on diverse safflower collections evaluated across different environments29,56.
Association analysis for seed yield identified distinct marker-trait associations (MTAs) in both environments, located on chromosomes 1, 2, 4, 6, 7 and 11. Among these, one MTA, Cart-SNP212, on chromosome 1, was share between both environments (Table 5). Previous studies have reported several markers associated with seed yield in safflower under drought conditions on chromosomes 6, 9 and 1229,58.
Additionally, one MTA (Cart-SNP4990) for 1000-seed weight was identified on chromosome 9, two MTAs, Cart-SNP2113 and Cart-SNP3276, located on chromosomes 4 and 6, respectively, were associated with the number of heads per plant, while another MTA (Cart-SNP2139) on chromosome 4 was associated with day to maturity. These MTAs were shared across both rainfed and supplementary irrigation environments. These findings provide valuable genetic resources for breeders aiming to improve desirable traits in safflower under diverse environmental conditions29. In our study, the locus Cart-SNP2549 was associated with relative water content (RWC) in the irrigated environment and with plant height (PH) in the rainfed environment, indicating a pleiotropic effect. This suggests linkage to different QTLs influencing both traits. Furthermore, Shared major genes or QTLs for different traits, such as day to flowering (DTF) and plant height (PH), have been previously reported in safflower29.
We observed site-specific MTAs for all measured traits, likely due to significant genotype-by-environment (G × E) interactions and variations in gene expression levels across different environments29,32. Although the QTLs identified for most traits in this study were located on chromosomes previously reported in the literature, we were unable to confirm the exact positions of the MTAs compared to those earlier findings.
This discrepancy may stem from differences in the safflower germplasm, the type of molecular markers used, and the distinct climatic conditions of the field evaluation sites. Given the complex nature of drought stress tolerance in plants, relying solely on direct yield and morphological traits may not be sufficient to identify QTLs associated with drought tolerance effectively.
In this study, GWAS analysis using STI values identified 45 significant MTAS, 12 of which overlapped with significant MTAs for morphological traits across both environments. To further investigate the potential gene functions of these stable MTAs, we performed BLAST searches within a 10-kb flanking region around the markers. Most of the significant SNP markers identified in this study aligned with candidate genes implicated in responses to abiotic stress in closely related species of Cathamus tinctorius. For instance, CCCH zinc-finger proteins are known to play crucial roles in plant development and responses to abiotic stresses such as salt, drought, flooding, low temperatures and oxidative stress. Similarly, candidate genes such as serine/threonine-protein kinase and dehydrogenase have been associated with drought-tolerance in plants59. Furthermore, the diacylglycerol acyltransferase gene is recognized for its role in wax ester biosynthesis and osmotic stress responses60.
Conclusion
The genetic dissection of morphological traits, their genotype-by-environment (G × E) interactions, and the identification of QTLs associated with drought tolerance are essential for developing climate-resilient safflower varieties. In this study, 90 globally diverse safflower genotypes were evaluated for seed yield, seed oil content, and morphological traits under two water regimes. The performance of these traits was significantly influenced by both rainfed and supplementary irrigation environments. Correlation analysis and PCA-biplot analysis highlighted traits such as the number of heads per plant (NHP), number of seeds per head (NSH), 1000-seed weight (TSW), and seed yield (Yp and Ys) as effective indicators for assessing diversity and characterizing drought-tolerant safflower genotypes. GWAS analysis based on morphological data from both environments and on STI values identified 66 and 45 significant marker-trait associations (MTAs), respectively. The environment-specific MTAs underscored the substantial impact of G × E interactions on all measured traits. Notably, stable MTAs across environments and those associated with STI values revealed several candidate genes with potential roles in abiotic stress response, providing a solid foundation for molecular breeding efforts. The newly identified MTAs and their chromosomal locations enhance our understanding of the genetic architecture underlying drought tolerance in safflower, paving the way for marker-assisted breeding to develop varieties with improved drought resilience.
Data availability
The data generated and/or analyzed during the current study are publicly available for legal/ethical reasons and also available from the corresponding author on reasonable request.
References
Talebi, R., Nosrati, S., Etminan, A. & Naji, A. M. Genetic diversity and population structure analysis of landrace and improved safflower (Cartamus tinctorious L.) germplasm using arbitrary functional gene-based molecular markers. Biotechnol. Biotechnol. Equip. 32, 1183–1194 (2018).
Joshi, S. et al. High-throughput phenotyping to dissect genotypic differences in safflower for drought tolerance. PLoS ONE 16(7), e0254908 (2021).
Talebi, R. & Abhari, S. A. Evaluation of genetic diversity in safflower (Carthamus tinctorius L.) using agro-morphological, fatty acid composition and ISSR molecular markers. Res. J. Biotechnol. 11(7), 19–27 (2016).
Hassani, S. M. R., Talebi, R., Pourdad, S. S., Naji, A. M. & Fayaz, F. Morphological description, genetic diversity and population structure of safflower (Carthamus tinctorius L.) mini core collection using SRAP and SSR markers. Biotechnol. Biotechnol. Equip. 34, 1043–1055 (2020).
Burke, J. M., Tang, S., Knapp, S. J. & Rieseberg, L. H. Genetic analysis of sunflower domestication. Genetics 161, 1257–1267 (2002).
Chapman, M. A., Hvala, J., Strever, J. & Burke, J. M. Population genetic analysis of safflower (Carthamus tinctorius; Asteraceae) reveals a Near Eastern origin and five centers of diversity. Am. J. Bot. 97, 831–840 (2010).
Hassani, S. M. R., Talebi, R., Pourdad, S. S., Naji, A. M. & Fayaz, F. In-depth genome diversity, population structure and linkage disequilibrium analysis of worldwide diverse safflower (Carthamus tinctorius L.) accessions using NGS data generated by DArTseq technology. Mol. Biol. Rep. 47(3), 2123–2135 (2020).
Nezhad, T. H. & Talebi, R. Interrelationships between agronomic traits with seed yield in safflower (Carthamus tinctorius L.) under different irrigation regimes. Biol. Forum Int. J. 7(2), 430–435 (2015).
Golkar, P., Hamzeh, E. & Mirmohammady Maibody, S. A. M. Discrimination of drought tolerance in a worldwide collection of safflower (Carthamus tinctorius L.) genotypes based on selection indices. Acta Agric. Slov. 117(1), 1–11 (2021).
Mahforojaki, M. S., Talebi, R. & Pourdad, S. S. Analysis of tolerance indices for germplasm evaluation in safflower (Carthamus tinctorius L.) subjected to terminal drought stress. South Asian J. Exp. Biol. 5(5), 156–161 (2015).
Yeloojeh, K. A. & Saeidi, G. Genetic analysis of safflower populations under water stress and non-stress conditions. J. Agron. 112, 3941–3947 (2020).
Kumar, S. et al. Utilization of molecular, phenotypic, and geographical diversity to develop compact composite core collection in the oilseed crop, safflower (Carthamus tinctorius L.) through maximization strategy. Front. Plant Sci. 7, 1554 (2016).
Hussain, M. I., Lyra, D. A., Farooq, M., Nikoloudakis, N. & Khalid, N. Salt and drought stresses in safflower: A review. Agron. Sustain. Dev. 36, 4 (2016).
Bailey-Serres, J., Parker, J. E., Ainsworth, E. A., Oldroyd, G. E. D. & Schroeder, J. I. Genetic strategies for improving crop yields. Nature 575(7781), 109–118 (2019).
Bao, X. et al. Screening and evaluation of drought resistance traits of winter wheat in the North China Plain. Front. Plant Sci. 14, 1194759 (2023).
Rabieyan, E. et al. Analysis of genetic diversity and genome-wide association study for drought tolerance related traits in Iranian bread wheat. BMC. Plant Biol. 23, 431 (2023).
Kumar, S. et al. Assessment of genetic diversity and population structure in a global reference collection of 531 accessions of Carthamus tinctorius L. (safflower) using AFLP markers. Plant Mol. Biol. Rep. 33, 1299–1313 (2015).
Bowers, J. E., Pearl, S. A. & Burke, J. M. Genetic mapping of millions of SNPs in safflower (Carthamus tinctorius L.) via whole genome resequencing. G3 6, 2203–2211 (2016).
Zhao, H. et al. Genomic prediction and genomic heritability of grain yield and its related traits in a safflower genebank collection. Plant Genome 14, e20064 (2021).
Wu, Z. et al. The chromosome-scale reference genome of safflower (Carthamus tinctorius) provides insights into linoleic acid and flavonoid biosynthesis. Plant Biotechnol. J. 19(9), 1725–1742 (2021).
Seyedimoradi, H., Talebi, R., Kanouni, H., Naji, A. M. & Karami, E. Genetic diversity and population structure analysis of chickpea (Cicer arietinum L.) advanced breeding lines using whole-genome DArTseq-generated SilicoDArT markers. Braz. J. Bot. 43, 541–549 (2020).
Gelaw, Y. M. et al. High-density DArTSeq SNP markers revealed wide genetic diversity and structured population in common bean (Phaseolus vulgaris L.) germplasm in Ethiopia. Mol. Biol. Rep. 50, 6739–6751 (2023).
Mujyambere, V., Adomako, K. & Olympio, O. S. Effectiveness of DArTseq markers application in genetic diversity and population structure of indigenous chickens in Eastern Province of Rwanda. BMC Genom. 25, 193 (2024).
Ali, F. et al. Identification of genetic basis associated with agronomic traits in a global safflower panel using genome wide association study. Turk. J. Agric. For. 45, 834–849 (2021).
Allan, V. et al. Genome-wide DArTSeq genotyping and phenotypic based assessment of within and among accessions diversity and effective sample size in the diverse sorghum, pearl millet, and pigeon pea landraces. Front. Plant Sci. 11, 587426 (2020).
Ibrahim, A. K. et al. Principles and approaches of association mapping in plant breeding. Trop. Plant Biol. 13, 212–224 (2020).
Yu, J. et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203–208 (2006).
Kaler, A. S., Gillman, J. D., Beissinger, T. & Purcell, L. C. Comparing different statistical models and multiple testing corrections for association mapping in soybean and maize. Front. Plant Sci. 10, 1794 (2020).
Zhao, H. et al. Genome-wide association studies dissect the G × E interaction for agronomic traits in a worldwide collection of safflowers (Carthamus tinctorius L.). Mol. Breed. 42, 24 (2022).
Zemour, K. et al. Phenol content and antioxidant and antiaging activity of safflower seed oil (Carthamus Tinctorius L.). Cosmetics 6, 55 (2019).
Turner, N. C. Techniques and experimental approaches for the measurement of plant water status. Plant Soil 58(1–3), 339–366 (1981).
Li, X., Guo, T., Mu, Q., Li, X. & Yu, J. Genomic and environmental determinants and their interplay underlying phenotypic plasticity. Proc. Natl. Acad Sci. 115, 6679 (2018).
Fernandez, G.C.J. Effective selection criteria for assessing plant stress tolerance. Proceeding Of The International Symposium On Adaptation Of Vegetables And Other Food Crops İn Temperature And Water Stress, Aug. 13–16, Shanhua, Taiwan, pp. 257–270 (1992).
Lassner, M. W., Peterson, P. & Yoder, J. I. Simultaneous amplification of multiple DNA fragments by polymerase chain reaction in the analysis of transgenic plants and their progeny. Plant Mol. Biol. Rep 7(2), 116–128 (1989).
Talebi, R. An alternative strategy in rapid DNA extraction protocol for high throughput RAPD analysis in chickpea and its wild related species. J. Appl. Biol. Sci. 2(3), 121–124 (2008).
Farahani, S. et al. Genome-wide association mapping for isolate-specific resistance to ascochyta rabiei in chickpea (Cicer arietinum L.). Physiol. Mol. Plant Pathol. 121, 101883 (2022).
Tomkowiak, A., Bocianowska, M. & Sobiech, A. The use of DArTseq technology to identify markers related to the heterosis effects in selected traits in maize. Curr. Issues Mol. Biol. 45, 2644–2660 (2023).
Talebi, R., Mahboubi, M., Naji, A. M. & Mehrabi, R. Physiological specialization of Puccinia triticina and genome-wide association mapping provide insights into the genetics of wheat leaf rust resistance in Iran. Sci. Rep. 13, 4398 (2023).
Mahboubi, M. et al. Genetic analysis of novel resistance sources and genome-wide association mapping identified novel QTLs for resistance to Zymoseptoria tritici, the causal agent of septoria tritici blotch in wheat. J. Appl. Genet. 63, 429–445 (2022).
Bradbury, P. J. et al. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635 (2007).
Perrier, X. & Jacquemoud-Collet, J. DARwin software http://darwin.cirad.fr/darwin (2006) .
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14, 2611–2620 (2005).
Earl, D. A. & Vonholdt, B. M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTU RE output and implementing the Evanno method. Conserv. Genet. Resour. 4(2), 359–361 (2012).
Peakall, R. & Smouse, P. E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539 (2012).
Shin, J. H., Blay, S., McNeney, B. & Graham, J. LDheatmap: An R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Softw. 16(3), 1–10 (2006).
Lipka, A. E. et al. GAPIT: Genome association and prediction integrated tool. Bioinformatics 28(18), 2397–2399 (2012).
Liu, X., Huang, M., Fan, B., Buckler, E. S. & Zhang, Z. Iterative usage of fxed and random efect models for powerful and efcient genome-wide association studies. PLoS Genet. 12, e1005767 (2016).
Huang, M., Liu, X., Zhou, Y., Summers, R. M. & Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. GigaScience 8(2), giy154 (2019).
Segura, V. et al. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 44, 825–830 (2012).
Çulha Erdal, Ş, Eyidoğan, F. & Ekmekçi, Y. Comparative physiological and proteomic analysis of cultivated and wild safflower response to drought stress and re-watering. Physiol. Mol. Biol. Plants 27(2), 281–295 (2021).
Yaghobnezhad, A., Talebi, R. & Pasari, B. The effect of foliar application of yeast and seaweed extract on the yield and quality of safflower seeds (Carthamus tinctorius) under drought stress condition. Iran. J. Plant Biotech. 18(1), 47–58 (2023).
Abraha, M. T., Shimelis, H., Laing, M. & Assefa, K. Gene action controlling yield and yield-related traits among tef (Eragrostis tet [Zucc] Trotter) populations under drought-stressed and non-stressed conditions. Plant Breed. 137, 585–597 (2018).
Mosupiemang, M., Emongor, V. E., Malambane, G. & Mapitse, R. Growth, development and yield of safflower genotypes in response to environmental variations. J. Phytol. 15, 45–154 (2023).
El-Hendawy, S. E., Hassan, W. M., Al-Suhaibani, N. A. & Schmidhalter, U. Spectral assessment of drought tolerance indices and grain yield in advanced spring wheat lines grown under full and limited water irrigation. Agric. Water Manag. 182(7), 1–12 (2017).
Balko, C., Torres, A. M. & Gutierrez, N. Variability in drought stress response in a panel of 100 faba bean genotypes. Front. Plant Sci. 14, 1236147 (2023).
Zhao, H., Lin, Z., Khansefid, M., Tibbits, J. F. & Hayden, M. J. Genomic prediction and selection response for grain yield in safflower. Front. Genet. 14, 1129433 (2023).
Timpson, N. J., Greenwood, C. M. T., Soranzo, N., Lawson, D. J. & Richards, J. B. Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat. Rev Genet. 19, 110–124 (2018).
Ebrahimi, F., Majidi, M. M., Arzani, A. & Mohammadi-Nejad, G. Association analysis of molecular markers with traits under drought stress in safflower. Crop Pasture Sci. 68, 167–175 (2017).
Lim, J., Lim, C. W. & Lee, S. C. Pepper novel serine-threonine kinase CaDIK1 regulates drought tolerance via modulating ABA sensitivity. Front. Plant Sci. 11, 1133 (2020).
Zhang, C., Yang, J., Meng, W., Zeng, L. & Sun, L. Genome-wide analysis of the WSD family in sunflower and functional identification of HaWSD9 involvement in wax ester biosynthesis and osmotic stress. Front. Plant Sci. 13, 975853 (2022).
Acknowledgements
The authors acknowledge the Deputy of Dryland Agricultural Research Institute Iran (Prof. Saeid Pourdad) for his kind support in germplasm preparation and support in field experiment.
Author information
Authors and Affiliations
Contributions
Seyed Mohammad Reza Hassani: Writing – original draft; Sayyed Saeid Pourdad: resources-supervision; Amir Mohammad Naji: Data curation, Formal analysis, Supervision; Farzad Fayaz: Data curation, Formal analysis; Seyed Mostafa Pirseyedi: Data curation, Formal analysis, review & editing; Siavash Sheikhizadeh Anari: Bioinformatics, Formal analysis; Reza Talebi: Conceptualization, Methodology, Resources, Writing – review & editing, Supervision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All the plant materials provided by Iranian Seed and Plant Improvement Institute (SPII) and Dryland Agricultural Research Institute (DARI) that were in compliance with relevant institutional, national, and international guidelines and legislation.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hassani, S.M.R., Pourdad, S.S., Naji, A.M. et al. Genome-wide association mapping in safflower (Carthamus tinctorius L.) for genetic dissection of drought tolerance using DArTseq markers. Sci Rep 14, 31490 (2024). https://doi.org/10.1038/s41598-024-82932-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-82932-4